Missense splice variant (g.20746A>G, p.Ile183Val) of interferon gamma receptor 1 (IFNGR1) coincidental with mycobacterial osteomyelitis - a screen of osteoarticular lesions
DOI:
https://doi.org/10.17305/bjbms.2016.1232Keywords:
Bacilli strain Calmette-Guerin, environmental mycobacteria, interferon gamma receptor 1, Mendelian susceptibility to mycobacterial disease, Mycobacterium bovis bacillus Calmette-Guérin, osteoarticular lesionsAbstract
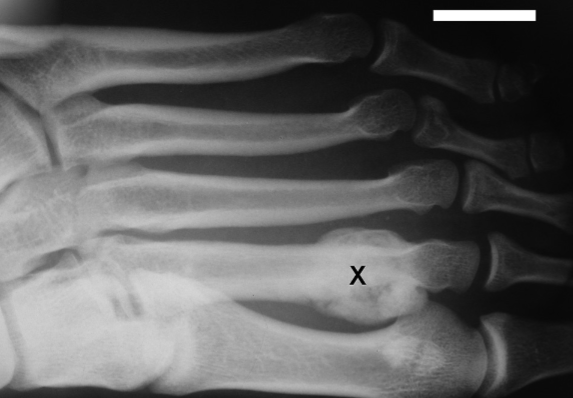
Previously, dominant partial interferon-gamma receptor 1 (IFN-g-R1) susceptibility to environmental mycobacteria was found with IFNGR1 deletions or premature stop. Our aim was to search for IFNGR1 variants in patients with mycobacterial osteoarticular lesions. Biopsies from the patients were examined for acid-fast bacilli, inflammatory cell infiltration, and mycobacterial niacin. Mycobacterial rRNA was analyzed using a target-amplified rRNA probe test. Peripheral-blood-leukocyte genomic DNA was isolated from 19 patients using the QIAamp DNA Mini Kit, and all IFNGR1 exons were sequenced using an ABIPRISM 3130 device. After the discovery of an exon 5 variant, a Polish newborn population sample (n = 100) was assayed for the discovered variant. Splice sites and putative amino acid interactions were analyzed. All patients tested were positive for mycobacteria; one was heterozygous for the IFNGR1 exon 5 single-nucleotide-missense substitution (g.20746A>G, p.Ile183Val). No other variant was found. The splice analysis indicated the creation of an exonic splicing silencer, and alternatively, molecular graphics indicated that the p.Ile183Val might alter beta-strand packing (loss of van der Waals contacts; Val183/Pro205), possibly altering the IFN-g-R1/IFN-g-R2 interaction. The probability of non-deleterious variant was estimated as <10%. Heterozygous IFNGR1:p.Ile183Val (frequency 0.003%) was found to be coincidental with mycobacterial osteomyelitis. The small amount of variation detected in the patients with osteoarticular lesions indicates that screens should not yet be restricted: Intronic variants should be analyzed as well as the other genes affecting Type 1 T-helper-cell-mediated immunity.
Citations
Downloads
References
. Cottle LE. Mendelian susceptibility to mycobacterial disease. Clin Genet 2011;79(1):17-22. http://dx.doi.org/10.1111/j.1399-0004.2010.01510.x.
. Bustamante J, Boisson-Dupuis S, Abel L, Casanova JL. Mendelian susceptibility to mycobacterial disease: Genetic, immunological, and clinical features of inborn errors of IFN-γ immunity. Semin Immunol 2014;26(6):454-70. http://dx.doi.org/10.1016/j.smim.2014.09.008.
. Thye T, Burchard GD, Nilius M, Müller-Myhsok B, Horstmann RD. Genomewide linkage analysis identifies polymorphism in the human interferon-gamma receptor affecting Helicobacter pylori infection. Am J Hum Genet 2003;72(2):448-53. http://dx.doi.org/10.1086/367714.
. Dorman SE, Picard C, Lammas D, Heyne K, van Dissel JT, Baretto R, et al. Clinical features of dominant and recessive interferon gamma receptor 1 deficiencies. Lancet 2004;364(9451):2113-21. http://dx.doi.org/10.1016/S0140-6736(04)17552-1.
. Tesi B, Sieni E, Neves C, Romano F, Cetica V, Cordeiro AI, et al. Hemophagocytic lymphohistiocytosis in 2 patients with underlying IFN-[gamma] receptor deficiency. J Allergy Clin Immunol 2015;135(6):1638. http://dx.doi.org/10.1016/j.jaci.2014.11.030.
. Aoki M, Matsui E, Kaneko H, Inoue R, Fukao T, Watanabe M, et al. A novel single-nucleotide substitution, Leu 467 Pro, in the interferon-gamma receptor 1 gene associated with allergic diseases. Int J Mol Med 2003;12(2):185-91. DOI: 10.3892/ijmm.12.2.185
. Fraser DA, Bulat-Kardum L, Knezevic J, Babarovic P, Matakovic-Mileusnic N, Dellacasagrande J, et al. Interferon-gamma receptor-1 gene polymorphism in tuberculosis patients from Croatia. Scand J Immunol 2003;57(5):480-4. http://dx.doi.org/10.1046/j.1365-3083.2003.01253.x.
. Matsuda A, Ebihara N, Kumagai N, Fukuda K, Ebe K, Hirano K, et al. Genetic polymorphisms in the promoter of the interferon gamma receptor 1 gene are associated with atopic cataracts. Invest Ophthalmol Vis Sci 2007;48(2):583-9. http://dx.doi.org/10.1167/iovs.06-0991.
. Cooke GS, Campbell SJ, Sillah J, Gustafson P, Bah B, Sirugo G, et al. Polymorphism within the interferon-γ/receptor complex is associated with pulmonary tuberculosis. Am J Respir Crit Care Med 2006;174(3):339-43. http://dx.doi.org/10.1164/rccm.200601-088OC.
. Zhou J, Chen DQ, Poon VK, Zeng Y, Ng F, Lu L, et al. A regulatory polymorphism in interferon-γ receptor 1 promoter is associated with the susceptibility to chronic hepatitis B virus infection. Immunogenetic 2009;61(6):423-30. http://dx.doi.org/10.1007/s00251-009-0377-8.
. Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res 2015;43:D470-8. http://dx.doi.org/10.1093/nar/gku1204.
. Tanaka Y, Nakashima H, Hisano C, Kohsaka T, Nemoto Y, Niiro H, et al. Association of the interferon-γ receptor variant (Val14Met) with systemic lupus erythematosus. Immunogenetics 1999;49(4):266-71. http://dx.doi.org/10.1007/s002510050492.
. Haverkamp MH, van de Vosse E, van Dissel JT. Nontuberculous mycobacterial infections in children with inborn errors of the immune system. J Infect 2014;68(Suppl 1):S134-50. http://dx.doi.org/10.1016/j.jinf.2013.09.024.
. Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, et al. Big data: Astronomical or genomical? PLoS Biol 2015;13(7):e1002195. http://dx.doi.org/10.1371/journal.pbio.1002195.
. van de Wetering D, de Paus RA, van Dissel JT, van de Vosse E. Functional analysis of naturally occurring amino acid substitutions in human IFN-gammaR1. Mol Immunol 2010;47(5):1023-30. http://dx.doi.org/10.1016/j.molimm.2009.11.016.
. Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: The next generation in gene variant databases. Hum Mutat 2011;32(5):557-63. http://dx.doi.org/10.1002/humu.21438.
. Loniewska B, Clark JS, Kaczmarczyk M, Adler G, Biñczak-Kuleta A, Kordek A, et al. Possible counter effect in newborns of 1936A>G (I646V) polymorphism in the AKAP10 gene encoding A-kinase-anchoring protein 10. J Perinatol 2012;32(3):230-4. http://dx.doi.org/10.1038/jp.2011.85.
. UniProt Consortium. UniProt: A hub for protein information. Nucleic Acids Res 2015;43:D204-12. DOI: 10.1093/nar/gku989.
. Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, et al. The UCSC genome browser database: 2014 update. Nucleic Acids Res 2014;42:D764-70. DOI: 10.1093/nar/gkt1168.
. SNPs S. NHLBI Program for Genomic Applications. Vol. 564. Seattle, WA: UW-FHCRC; 2007. p. 565-6. Available from: http://www.pga.gs.washington.edu. [Last accessed on 31 Aug 2015].
. Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 2014;42:D980-5. DOI: 10.1093/nar/gkt1113.
. Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, et al. Ensembl genomes 2016: More genomes, more complexity. Nucleic Acids Res 2016;44(D1):D574-80. DOI: 10.1093/nar/gkv1209
. DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA, USA: Scientific; 2002.
. Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human splicing finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res 2009;37(9):e67. DOI: 10.1093/nar/gkp215.
. Lee SB, Kim BC, Jin SH, Park YG, Kim SK, Kang TJ, et al. Missense mutations of the interleukin-12 receptor beta 1(IL12RB1) and interferon-gamma receptor 1 (IFNGR1) genes are not associated with susceptibility to lepromatous leprosy in Korea. Immunogenetics 2003;55(3):177-81.
http://dx.doi.org/10.1007/s00251-003-0573-x.
. Quispel WT, Stegehuis-Kamp JA, Santos SJ, van Wengen A, Dompeling E, Egeler RM, et al. Intact IFN-γR1 expression and function distinguishes langerhans cell histiocytosis from Mendelian susceptibility to mycobacterial disease. J Clin Immunol 2014;34(1):84-93. http://dx.doi.org/10.1007/s10875-013-9959-1.
. Fieschi C, Dupuis S, Picard C, Smith CE, Holland SM, Casanova JL. High levels of interferon gamma in the plasma of children with complete interferon gamma receptor deficiency. Pediatrics 2001;107(4):E48.
. Kong XF, Vogt G, Chapgier A, Lamaze C, Bustamante J, Prando C, et al. A novel form of cell type-specific partial IFN-gammaR1 deficiency caused by a germ line mutation of the IFNGR1 initiation codon. Hum Mol Genet 2010;19(3):434-44. http://dx.doi.org/10.1093/hmg/ddp507.
. Noordzij JG, Hartwig NG, Verreck FA, De Bruin-Versteeg S, De Boer T, Van Dissel JT, et al. Two patients with complete defects in interferon gamma receptor-dependent signaling. J Clin Immunol 2007;27(5):490-6.
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2016 Bosnian Journal of Basic Medical Sciences

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2016-05-05
Published 2016-08-02









