Small interfering RNA-mediated silencing of nicotinamide phosphoribosyltransferase (NAMPT) and lysosomal trafficking regulator (LYST) induce growth inhibition and apoptosis in human multiple myeloma cells: A preliminary study
DOI:
https://doi.org/10.17305/bjbms.2016.1568Keywords:
Multiple myeloma, nicotinamide phosphoribosyltransferase, lysosomal trafficking regulator, small interfering RNA, cell proliferation, apoptosisAbstract
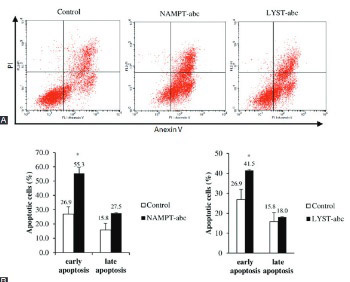
Multiple myeloma (MM) is a malignancy of B lymphocytes or plasma cells. Our array-based comparative genomic hybridization findings revealed chromosomal gains at 7q22.3 and 1q42.3, where nicotinamide (NAM) phosphoribosyltransferase (NAMPT) and lysosomal trafficking regulator (LYST) genes are localized, respectively. This led us to further study the functions of these genes in myeloma cells. NAMPT is a key enzyme involved in nicotinamide adenine dinucleotide salvage pathway, and it is frequently overexpressed in human cancers. In contrast, little is known about the function of LYST in cancer. The expression of LYST is shown to affect lysosomal size, granule size, and autophagy in human cells. In this study, the effects of small interfering RNA (siRNA)-mediated silencing of NAMPT and LYST on cell proliferation and apoptosis were evaluated in RPMI 8226 myeloma cells. Transfection efficiencies were determined by quantitative real time reverse transcriptase PCR. Cell proliferation was determined using MTT assay, while apoptosis was analyzed with flow cytometry using Annexin V-fluorescein isothiocyanate/propidium iodide assay. The NAMPT protein expression in siRNA-treated cells was estimated by enzyme-linked immunosorbent assay. Our results showed that NAMPT and LYST were successfully knockdown by siRNA transfection (p < 0.05). NAMPT or LYST gene silencing significantly inhibited cell proliferation and induced apoptosis in RPMI 8226 cells (p < 0.05). Silencing of NAMPT gene also decreased NAMPT protein levels (p < 0.01). Our study demonstrated that NAMPT and LYST play pivotal roles in the molecular pathogenesis of MM. This is the first report describing the possible functions of LYST in myelomagenesis and its potential role as a therapeutic target in MM.
Citations
Downloads
References
Eslick R, Talaulikar D. Multiple myeloma: From diagnosis to treatment. Aust Fam Physician. 2013;42(10):684-8.
Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: The experience of the Intergroupe Francophone du Myélome. Blood. 2007;109(8):3489-95. http://dx.doi.org/10.1182/blood-2006-08-040410.
Omar ZA, Ibrahim Tamin NS. National cancer registry report: Malaysia cancer statistics-data and figure. Malaysia: National Cancer Registry; 2011. p. 85-7.
Sharma A, Heuck CJ, Fazzari MJ, Mehta J, Singhal S, Greally JM, et al. DNA methylation alterations in multiple myeloma as a model for epigenetic changes in cancer. Wiley Interdiscip Rev Syst Biol Med. 2010;2(6):654-69. http://dx.doi.org/10.1002/wsbm.89.
Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467-72. http://dx.doi.org/10.1038/nature09837.
Dimopoulos K, Gimsing P, Grønbæk K. The role of epigenetics in the biology of multiple myeloma. Blood Cancer J. 2014;4:e207. http://dx.doi.org/10.1038/bcj.2014.29.
Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279(49):50754-63. http://dx.doi.org/10.1074/jbc.M408388200.
Shackelford RE, Mayhall K, Maxwell NM, Kandil E, Coppola D. Nicotinamide phosphoribosyltransferase in malignancy: A review. Genes Cancer. 2013;4(11-12):447-56. http://dx.doi.org/10.1177/1947601913507576.
Chiarugi A, Dölle C, Felici R, Ziegler M. The NAD metabolome – A key determinant of cancer cell biology. Nat Rev Cancer. 2012;12(11):741-52. http://dx.doi.org/10.1038/nrc3340.
de la Puente P, Muz B, Azab F, Luderer M, Azab AK. Molecularly targeted therapies in multiple myeloma. Leuk Res Treatment. 2014;2014:976567.
DOI: 10.1155/2014/976567.
Prideaux SM, Conway O'Brien E, Chevassut TJ. The RAG model: A new paradigm for genetic risk stratification in multiple myeloma. Bone Marrow Res. 2014;2014:526568. http://dx.doi.org/10.1155/2014/526568.
Cullinane AR, Schäffer AA, Huizing M. The BEACH is hot: A LYST of emerging roles for BEACH-domain containing proteins in human disease. Traffic. 2013;14(7):749-66. http://dx.doi.org/10.1111/tra.12069.
Al-Tamemi S, Al-Zadjali S, Al-Ghafri F, Dennison D. Chediak-Higashi syndrome: Novel mutation of the CHS1/LYST gene in 3 Omani patients. J Pediatr Hematol Oncol. 2014;36(4):e248-50. http://dx.doi.org/10.1097/MPH.0000000000000025.
Jessen B, Maul-Pavicic A, Ufheil H, Vraetz T, Enders A, Lehmberg K, et al. Subtle differences in CTL cytotoxicity determine susceptibility to hemophagocytic lymphohistiocytosis in mice and humans with Chediak-Higashi syndrome. Blood. 2011;118(17):4620-9. http://dx.doi.org/10.1182/blood-2011-05-356113.
Sepulveda FE, Burgess A, Heiligenstein X, Goudin N, Ménager MM, Romao M, et al. LYST controls the biogenesis of the endosomal compartment required for secretory lysosome function. Traffic. 2015;16(2):191-203. http://dx.doi.org/10.1111/tra.12244.
Ivyna Bong PN, Ng CC, Lam KY, Megat Baharuddin PJ, Chang KM, Zakaria Z. Identification of novel pathogenic copy number aberrations in multiple myeloma: The Malaysian context. Mol Cytogenet. 2014;7(1):24.
http://dx.doi.org/10.1186/1755-8166-7-24.
Rongvaux A, Andris F, Van Gool F, Leo O. Reconstructing eukaryotic NAD metabolism. Bioessays. 2003;25(7):683-90. http://dx.doi.org/10.1002/bies.10297.
Zhang T, Berrocal JG, Frizzell KM, Gamble MJ, DuMond ME, Krishnakumar R, et al. Enzymes in the NAD salvage pathway regulate SIRT1 activity at target gene promoters. J Biol Chem. 2009;284(30):20408-17. http://dx.doi.org/10.1074/jbc.M109.016469.
Revollo JR, Körner A, Mills KF, Satoh A, Wang T, Garten A, et al. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007;6(5):363-75. http://dx.doi.org/10.1016/j.cmet.2007.09.003.
van der Veer E, Nong Z, O'Neil C, Urquhart B, Freeman D, Pickering JG. Pre-B-cell colony-enhancing factor regulates NAD - Dependent protein deacetylase activity and promotes vascular smooth muscle cell maturation. Circ Res. 2005;97(1):25-34. http://dx.doi.org/10.1161/01.RES.0000173298.38808.27.
van der Veer E, Ho C, O'Neil C, Barbosa N, Scott R, Cregan SP, et al. Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J Biol Chem. 2007;282(15):10841-5. http://dx.doi.org/10.1074/jbc.C700018200.
Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14(5):661-73. http://dx.doi.org/10.1016/j.devcel.2008.02.004.
Rongvaux A, Galli M, Denanglaire S, Van Gool F, Drèze PL, Szpirer C, et al. Nicotinamide phosphoribosyl transferase/pre-B cell colony-enhancing factor/visfatin is required for lymphocyte development and cellular resistance to genotoxic stress. J Immunol. 2008;181(7):4685-95. http://dx.doi.org/10.4049/jimmunol.181.7.4685.
Cea M, Cagnetta A, Fulciniti M, Tai YT, Hideshima T, Chauhan D, et al. Targeting NAD salvage pathway induces autophagy in multiple myeloma cells via mTORC1 and extracellular signal-regulated kinase (ERK1/2) inhibition. Blood. 2012;120(17):3519-29. http://dx.doi.org/10.1182/blood-2012-03-416776.
Kim MY, Woo EM, Chong YT, Homenko DR, Kraus WL. Acetylation of estrogen receptor alpha by p300 at lysines 266 and 268 enhances the deoxyribonucleic acid binding and transactivation activities of the receptor. Mol Endocrinol. 2006;20(7):1479-93. http://dx.doi.org/10.1210/me.2005-0531.
Michan S, Sinclair D. Sirtuins in mammals: Insights into their biological function. Biochem J. 2007;404(1):1-13. http://dx.doi.org/10.1042/BJ20070140.
Dvir-Ginzberg M, Gagarina V, Lee EJ, Hall DJ. Regulation of cartilage-specific gene expression in human chondrocytes by SirT1 and nicotinamide phosphoribosyltransferase. J Biol Chem. 2008;283(52):36300-10. http://dx.doi.org/10.1074/jbc.M803196200.
Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8(5):347-58. http://dx.doi.org/10.1016/j.cmet.2008.08.017.
Schiewer MJ, Goodwin JF, Han S, Brenner JC, Augello MA, Dean JL, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2(12):1134-49. http://dx.doi.org/10.1158/2159-8290.CD-12-0120.
Ko HL, Ren EC. Functional aspects of PARP1 in DNA repair and transcription. Biomolecules. 2012;2(4):524-48. http://dx.doi.org/10.3390/biom2040524.
Teoh PJ, Chng WJ. p53 abnormalities and potential therapeutic targeting in multiple myeloma. Biomed Res Int. 2014;2014:717919. DOI: 10.1155/2014/717919.
Abdi J, Chen G, Chang H. Drug resistance in multiple myeloma: Latest findings and new concepts on molecular mechanisms. Oncotarget. 2013;4(12):2186-207. http://dx.doi.org/10.18632/oncotarget.1497.
Zhou Y, Uddin S, Zimmerman T, Kang JA, Ulaszek J, Wickrema A. Growth control of multiple myeloma cells through inhibition of glycogen synthase kinase-3. Leuk Lymphoma. 2008;49(10):1945-53. http://dx.doi.org/10.1080/10428190802304966.
Hannus M, Beitzinger M, Engelmann JC, Weickert MT, Spang R, Hannus S, et al. siPools: Highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 2014;42(12):8049-61. http://dx.doi.org/10.1093/nar/gku480.
Cea M, Cagnetta A, Patrone F, Nencioni A, Gobbi M, Anderson KC. Intracellular NAD+ depletion induces autophagic death in multiple myeloma cells. Autophagy. 2013;9(3):410-2. http://dx.doi.org/10.4161/auto.22866.
Faigle W, Raposo G, Tenza D, Pinet V, Vogt AB, Kropshofer H, et al. Deficient peptide loading and MHC class II endosomal sorting in a human genetic immunodeficiency disease: The Chediak-Higashi syndrome. J Cell Biol. 1998;141(5):1121-34. http://dx.doi.org/10.1083/jcb.141.5.1121.
Downloads
Additional Files
Published
Issue
Section
Categories
How to Cite
Accepted 2016-08-20
Published 2016-11-10









