Tumor suppressive function of microRNA-192 in acute lymphoblastic leukemia
DOI:
https://doi.org/10.17305/bjbms.2017.1921Keywords:
Acute lymphoblastic leukemia, apoptosis, cell cycle, miroRNA-192, ALL, P53, BAX, CASP3, BCL-2Abstract
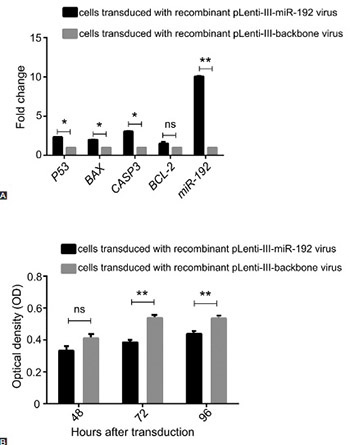
Non-coding RNAs play a critical role in gene regulation in cancer cells. Reduced expression of microRNA-192 (miR-192) has been detected in many cancers. In this study, we investigated the role of miR-192 in cell proliferation and cell cycle control in NALM-6 cell line, a model of acute lymphoblastic leukemia (ALL). Cell cycle analysis by DNA content using propidium iodide staining and cell apoptosis analysis using Annexin V assay were carried out. Cell proliferation changes were monitored using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. In addition, the relative changes in P53, BAX, CASP3, and BCL-2 gene expression were determined by quantitative reverse transcription PCR. Overexpression of miR-192 resulted in cell proliferation arrest in ALL cells. After 72 and 96 hours of transduction, apoptosis was significantly increased in the cells transduced with miR-192-overexpressing virus compared with control cells. The expression of P53, BAX, and CASP3 increased after 48 hours of transduction in miR-192-overexpressing cells, but no change was observed in BCL-2 expression. The G0/S and G1/S ratio changed to 7.5 and 4.5, respectively, in the cells overexpressing miR-192 compared with controls. The results of our study suggest, for the first time, tumor suppressive effects of miR-192 in ALL cells.
Citations
Downloads
References
Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116(2):281-97. https://doi.org/10.1016/S0092-8674(04)00045-5.
Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell 2009;136(2):215-33. https://doi.org/10.1016/j.cell.2009.01.002.
Ruvkun G. Clarifications on miRNA and cancer. Science 2006;311(5757):36-7. https://doi.org/10.1126/science.311.5757.36d.
Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol 2007;302(1):1-12. https://doi.org/10.1016/j.ydbio.2006.08.028.
Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res 2006;12(7 Pt 1):2014-24. https://doi.org/10.1158/1078-0432.CCR-05-1853.
Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science 2003;299(5612):1540. https://doi.org/10.1126/science.1080372.
Hino K, Tsuchiya K, Fukao T, Kiga K, Okamoto R, Kanai T, et al. Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. RNA 2008;14(7):1433-42. https://doi.org/10.1261/rna.810208.
Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X, et al. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS One 2014;9(9):e107986. https://doi.org/10.1371/journal.pone.0107986.
Feinberg-Gorenshtein G, Guedj A, Shichrur K, Jeison M, Luria D, Kodman Y, et al. MiR-192 directly binds and regulates Dicer1 expression in neuroblastoma. PLoS One 2013;8(11):e78713. https://doi.org/10.1371/journal.pone.0078713.
Jin Z, Selaru FM, Cheng Y, Kan T, Agarwal R, Mori Y, et al. MicroRNA-192 and -215 are upregulated in human gastric cancer in vivo and suppress ALCAM expression in vitro. Oncogene 2011;30(13):1577-85. https://doi.org/10.1038/onc.2010.534.
Song B, Wang Y, Kudo K, Gavin EJ, Xi Y, Ju J. miR-192 regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clin Cancer Res 2008;14(24):8080-6. https://doi.org/10.1158/1078-0432.CCR-08-1422.
Schotte D, De Menezes RX, Akbari Moqadam F, Khankahdani LM, Lange-Turenhout E, Chen C, et al. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica 2011;96(5):703-11. https://doi.org/10.3324/haematol.2010.026138.
Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol 2013;15(1):2-8. https://doi.org/10.1038/ncb2641.
Surget S, Khoury MP, Bourdon JC. Uncovering the role of p53 splice variants in human malignancy: A clinical perspective. Onco Targets Ther 2013;7:57-68. https://doi.org/10.2147/OTT.S53876.
Tudzarova S, Mulholland P, Dey A, Stoeber K, Okorokov AL, Williams GH, et al. p53 controls CDC7 levels to reinforce G1 cell cycle arrest upon genotoxic stress. Cell Cycle 2016;15(21):2958-72. https://doi.org/10.1080/15384101.2016.1231281.
Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene 2001;20(15):1803-15. https://doi.org/10.1038/sj.onc.1204252.
Khella HW, Bakhet M, Allo G, Jewett MA, Girgis AH, Latif A, et al. miR-192, miR-194 and miR-215: A convergent microRNA network suppressing tumor progression in renal cell carcinoma. Carcinogenesis 2013;34(10):2231-9. https://doi.org/10.1093/carcin/bgt184.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435(7043):834-8. https://doi.org/10.1038/nature03702.
Schwartz D, Almog N, Peled A, Goldfinger N, Rotter V. Role of wild type p53 in the G2 phase: Regulation of the gamma-irradiation-induced delay and DNA repair. Oncogene 1997;15(21):2597-607. https://doi.org/10.1038/sj.onc.1201436.
Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 1993;74(4):609-19. https://doi.org/10.1016/0092-8674(93)90509-O.
Weng C, Li Y, Xu D, Shi Y, Tang H. Specific cleavage of Mcl-1 by caspase-3 in tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in Jurkat leukemia T cells. J Biol Chem 2005;280(11):10491-500. https://doi.org/10.1074/jbc.M412819200.
Jin Y, Lu J, Wen J, Shen Y, Wen X. Regulation of growth of human bladder cancer by miR-192.Tumour Biol 2015;36(5):3791-7. DOI: 10.1007/s13277-014-3020-8.
Downloads
Additional Files
Published
Issue
Section
Categories
How to Cite
Accepted 2017-03-15
Published 2017-08-20









