Evaluation of emm gene types, toxin gene profiles and clonal relatedness of group A streptococci
DOI:
https://doi.org/10.17305/bjbms.2013.2356Keywords:
Group A streptococci, emm typing, toxin genes, speA, speB, speC, clonal relation, PFGEAbstract
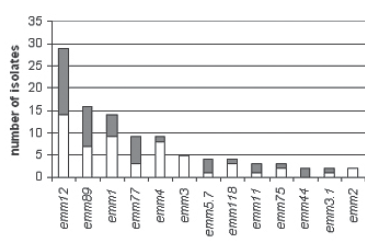
The aim of this study is to evaluate antibiotic susceptibilities, emm gene types, toxin gene profiles and clonal relatedness of group A streptococci (GAS) isolates obtained from patients and carriers. A total of 79 clinical isolates from patients and 60 isolates from carriers were included in the study. Emm typing, toxin gene detection for speA, speB, speC, speG and smeZ genes and pulsed-field gel electrophoresis (PFGE) was performed. Twenty-one distinct emm types were detected; the most common types were emm12, emm89, emm1, emm77, emm4 and emm3. The detection rates of both emm types and the toxingenes didn't differ significantly between patients and carriers. The presence of speA and smeZ was significantly higher in emm1 and speG was significantly lower in emm4 when compared to the other emm types. The rate of clustering obtained with PFGE wasn't significantly different in patients and carriers. As a result, twelve of the 21 emm types detected in this study were covered by the 26-valent vaccine, constituting 77.7% of the emm typeable isolates; however the emm4 type which is one of the most common types in the present study is not among this coverage.
Citations
Downloads
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2017 Bosnian Journal of Basic Medical Sciences

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2017-08-13
Published 2013-08-20









