Prevalence and genotype distribution of rotaviruses in children with gastroenteritis in Rize province
DOI:
https://doi.org/10.17305/bjbms.2015.469Keywords:
Rotaviruses, children, genotypingAbstract
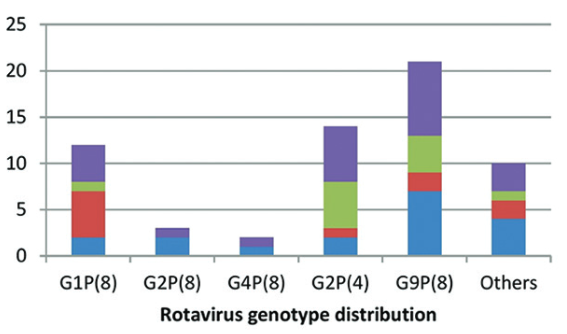
Determination of the distribution of rotavirus genotypes is essential for understanding the epidemiology of this virus responsible for nearly half a million of deaths in patients with gastroenteritis worldwide. In the present study, we aimed to genotype the rotavirus strains isolated from diarrheal stool samples in children under 5 years old. A total of 1297 fecal samples were collected, and rotavirus antigen was detected in 73 of these samples. Antigen-positive samples were transferred to the Public Health Agency of Turkey, Molecular Microbiology Research Laboratory, and were tested for determination of genotypes G and P using semi-nested multiplex polymerase chain reaction method performed with consensus- and genotype-specific primers. Twelve specimens were found to be negative for rotavirus in genotyping method. All the positive-strains were in G1-4, G8-9, P(4), P(8), and P(9) genotypes. The most frequent GP genotype combinations were found to be G9P(8) in 21 strains (34.4%), G2P(4) in 14 strains (23.0%), and G1P(8) in 12 strains (19.7%). We found 10 distinct genotypes amongst a total of 61 strains. Among the strains isolated and genotyped in our study, 90.2% (55/61) and 67.2% (41/61) have already been included in the two existing commercial vaccines. In conclusion, these findings implicate the necessity of development of region-specific vaccines after evaluation of the local genotype distribution. Further studies on the large number of rotavirus strains would contribute to this process.
Citations
Downloads
References
Kim MJ, Jeong HS, Kim SG, Lee SM, Kim SH, Kee HY, et al. Diversity of rotavirus strain circulated in gwangju, republic of Korea. Osong Public Health Res Perspect 2014;5(6):364–369. doi:10.1016/j.phrp.2014.10.004.
Matthijnssens J, Otto PH, Ciarlet M. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch Virol 2012;157(6):1177–1182. doi:10.1007/s00705-012-1273-3.
CDC. Prevention of rotavirus gastroenteritis among infants and children: recommendation of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 2009; 58(2):1–25.
Durmaz R, Kalaycioglu AT, Acar S, Bakkaloglu Z, Karagoz A, Korukluoglu G, et al. Turkish Rotavirus Surveillance Network. Prevalence of Rotavirus Genotypes in Children Younger than 5 Years of Age before the Introduction of a Universal Rotavirus Vaccination Program: Report of Rotavirus Surveillance in Turkey. PLoS One 2014;9(12):e113674. doi:10.1371/journal.pone.0113674.
Iturriza-Gómara M, Dallman T, Bányai K, Böttiger B, Buesa J, Diedrich S, et al. Rotavirus genotypes co-circulating in Europe between 2006 and 2009 as determined by EuroRotaNet, a pan-European collaborative strain surveillance network. Epidemiol Infect 2011;139(6):895-909. doi:10.1017/S0950268810001810.
Simmonds MK, Armah G, Asmah R, Banerjee I, Damanka S, Esona M, et al. New oligonucleotide primers for P-typing of rotavirus strains: Strategies for typing previously untypeable strains. J Clin Virol 2008;42(4):368–373.doi :10.1016/j.jcv.2008.02.011.
Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Banyai K, Ramachandran M, et al. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J Infect Dis 2005;192(1):146–159.doi :10.1086/431499.
Iturriza-Gómara M., Cubitt D., Desselberger U. Amino acid substitution within the VP7 protein of G2 rotavirus strains associated with failure to serotype. J Clin Microbiol 2001;39(10):3796–3798.doi :10.1128/JCM.39.10.3796-3798.2001.
Tapisiz A, Karahan ZC, Ciftci E, Ince E, Dogru U. Changing patterns of rotavirus genotypes in Turkey. Curr Microbiol 2011;63(6):517–522.doi:10.1007/s00284-011-0014-2.
Bozdayi G, Dogan B, Dalgic B, Bostanci I, Sari S, Battaloglu NO, et al. Diversity of human rotavirus G9 among children in Turkey. J Med Virol 2008;80(4):733–740.doi:10.1002/jmv.21120.
Midgley S, Böttiger B, Jensen TG, Friis-Møller A, Person LK, Nielsen L, et al. Human group A rotavirus infections in children in Denmark; detection of reassortant G9 strains and zoonotic P(14) strains. Infect Genet Evol 2014;27:114–120.doi:10.1016/j.meegid.2014.07.008.
Anca IA, Furtunescu FL, Pleţca D, Streinu-Cercel A, Rugină S, Holl K. Hospital-based surveillance to estimate the burden of rotavirus gastroenteritis in children below five years of age in Romania. Germs 2014;4(2):30–40.doi:10.11599/germs.2014.1053.
Mullick S, Mukherjee A, Ghosh S, Pazhani GP, Sur D, Manna B, et al. Community based case-control study of rotavirus gastroenteritis among young children during 2008-2010 reveals vast genetic diversity and increased prevalence of G9 strains in Kolkata. PLoS One 2014;17;9(11):e112970. doi:10.1371/journal.pone.0112970.
Numazaki K, Ichikawa M. Clinical and virological characteristics of rotavirus gastroenteritis and prevalence of strains in Tochigi, Japan. In Vivo 2014;28(6):1141–1147.
Boula A, Waku-Kouomou D, Njiki Kinkela M, Esona MD, Kemajou G, Mekontso D, et al. Molecular surveillance of rotavirus strains circulating in Yaoundé, Cameroon, September 2007-December 2012. Infect Genet Evol 2014;28:470–475.doi:10.1016/j.meegid.2014.08.019.
Semeiko GV, Yermalovich MA, Poliakova N, Mijatovic-Rustempasic S, Kerin TK, Wasley A, et al. Rotavirus genotypes in Belarus, 2008-2012. Infect Genet Evol 2014;28:480–485.doi:10.1016/j.meegid.2014.09.007.
Cataloluk O, Iturriza M, Gray J. Molecular characterization of rotaviruses circulating in the population of Turkey. Epidemiol Infect 2005;133(4):673–678.doi:10.1017/S0950268805003882.
Altindis M, Bányai K, Kalayci R, Gulamber C, Koken R, Apan T, et al. Molecular characterization of rotaviruses in mid-western Turkey, 2006–2007. Cent Eur J Med 2010;5(5):640–645.doi :10.2478/s11536-009-0130-6.
Ruiz-Palacios GM, Pérez-Schael I, Velázquez FR, Abate H, Breuer T, Clemens SC et al. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med 2006;354:11–22.doi:10.1056/NEJMoa052434.
Dulgheroff AC, Figueiredo EF, Gouvêa VS, Domingues AL. Changes in epidemiology of rotavirus in the Triângulo Mineiro region of Brazil: lack of two consecutive rotavirus seasons. Braz J Med Biol Res 2014;47(12):1091–1095.doi:10.1590/1414-431X20144156.
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2015 Bosnian Journal of Basic Medical Sciences

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2015-04-29
Published 2015-07-09









