Detailed polymorphism study on cytomegalovirus DNA polymerase gene to reveal the most suitable genomic targets for quantitative Real-time PCR
DOI:
https://doi.org/10.17305/bjbms.2015.494Keywords:
Cytomegalovirus, Real-time PCR, Nucleotide variationsAbstract
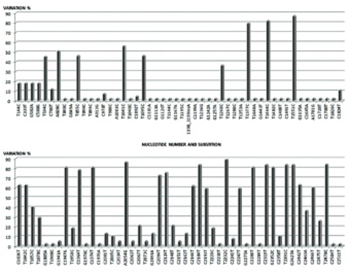
The human cytomegalovirus (HCMV) is an important human pathogen primarily affecting immunocompromised patients, like transplant recipients or HIV- infected individuals. Early diagnosis of cytomegalovirus (CMV) infection in high-risk patients is essential in order to start preemptive treatments. pol (UL54) gene encoding for HCMV viral DNA polymerase is a well-defined target for HCMV detection in clinical samples and identifying most highly conserved regions for primer design remains crucial. Though real-time polymerase chain reaction (qPCR) is a rapid and sensitive method for HCMV detection, failure to detect some HCMV strains due to primer and target mismatches have led the researchers to explore more sensitive and reliable methods. Hence, to understand the broader diversity of the pol mutations in HCMV and to specify the most suitable region for primer-probe design to be used in qPCR assay, we studied both nucleotide and amino acid heterogeneities in 60 HCMV positive samples that were collected to represent national mutational prevalence of pol gene of HCMV in Turkey. The test was designed with a new set of primers- probe for HCMV detection and quantification based on the sequencing data which revealed the most conserved region on the pol gene. Statistical probit analysis was applied on qPCR studies which revealed a 95% detection limit of 100 copies/mL. In addition, linearity, reproducibility, and precision of the new test were assessed for diagnostic purposes.
Citations
Downloads
References
Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78-83.
Chou SW. Acquisition of donor strains of cytomegalovirus by renal-transplant recipients. N Engl J Med. 1986;314:1418-23. http://dx.doi.org/10.1056/NEJM198605293142205.
Dollard SC, Grosse SD, Ross DS. New estimates of the prevalence of neurological and sensory sequelae and mortality associated with congenital cytomegalovirus infection. Rev Med Virol. 2007;17:355-63. http://dx.doi.org/10.1002/rmv.544.
Torres-Madriz G, Boucher HW. Immunocompromised hosts: Perspectives in the treatment and prophylaxis of cytomegalovirus disease in solid-organ transplant recipients. Clin Infect Dis. 2008;47:702-11. http://dx.doi.org/10.1086/590934.
Chrisp P, Clissold SP. Foscarnet. A review of its antiviral activity, pharmacokinetic properties and therapeutic use in immunocompromised patients with cytomegalovirus retinitis. Drugs. 1991;41:104-29. http://dx.doi.org/10.2165/00003495-199141010-00009.
Lalezari JP. Cidofovir: A new therapy for cytomegalovirus retinitis. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;14:S22-6. http://dx.doi.org/10.1097/00042560-199700001-00005, http://dx.doi.org/10.1097/00042560-199700001-00006.
Pescovitz MD, Rabkin J, Merion RM, Paya CV, Pirsch J, Freeman RB, et al. Valganciclovir results in improved oral absorption of ganciclovir in liver transplant recipients. Antimicrob Agents Chemother. 2000;44:2811-5. http://dx.doi.org/10.1128/AAC.44.10.2811-2815.2000.
Sullivan V, Talarico CL, Stanat SC, Davis M, Coen DM, Biron KK. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature. 1992;358:162-4. http://dx.doi.org/10.1038/358162a0.
Baldanti F, Underwood MR, Talarico CL, Simoncini L, Sarasini A, Biron KK, et al. The Cys607-->Tyr change in the UL97 phosphotransferase confers ganciclovir resistance to two human cytomegalovirus strains recovered from two immunocompromised patients. Antimicrob Agents Chemother. 1998;42:444-6.
Kotton CN. CMV: Prevention, diagnosis and therapy. Am J Transplant. 2013;13:24-40. http://dx.doi.org/10.1111/ajt.12006.
Zweygberg Wirgart B, Brytting M, Linde A, Wahren B, Grillner L. Sequence variation within three important cytomegalovirus gene regions in isolates from four different patient populations. J Clin Microbiol. 1998;36:3662-9.
Erice A. Resistance of human cytomegalovirus to antiviral drugs. Clin Microbiol Rev. 1999;12:286-97.
Weinberg A, Jabs DA, Chou S, Martin BK, Lurain NS, Forman MS, et al. Mutations conferring foscarnet resistance in a cohort of patients with acquired immunodeficiency syndrome and cytomegalovirus retinitis. J Infect Dis. 2003;187:777-84. http://dx.doi.org/10.1086/368385.
Gilbert C, Boivin G. Human cytomegalovirus resistance to antiviral drugs. Antimicrob Agents Chemother. 2005;49:873-83. http://dx.doi.org/10.1128/AAC.49.3.873-883.2005.
Chou S, Lurain NS, Thompson KD, Miner RC, Drew WL. Viral DNA polymerase mutations associated with drug resistance in human cytomegalovirus. J Infect Dis. 2003;188:32-9. http://dx.doi.org/10.1086/375743.
Chou SW, Dennison KM. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J Infect Dis. 1991;163:1229-34. http://dx.doi.org/10.1093/infdis/163.6.1229.
Fox JC, Griffiths PD, Emery VC. Quantification of human cytomegalovirus DNA using the polymerase chain reaction. J Gen Virol. 1992;73:2405-8. http://dx.doi.org/10.1099/0022-1317-73-9-2405.
Guiver M, Fox AJ, Mutton K, Mogulkoc N, Egan J. Evaluation of CMV viral load using TaqMan CMV quantitative PCR and comparison with CMV antigenemia in heart and lung transplant recipients. Transplantation. 2001;71:1609-15. http://dx.doi.org/10.1097/00007890-200106150-00021.
Novak Z, Chowdhury N, Ross SA, Pati SK, Fowler K, Boppana SB. Diagnostic consequences of cytomegalovirus glycoprotein B polymorphisms. J Clin Microbiol. 2011;49:3033-5. http://dx.doi.org/10.1128/JCM.01039-11.
Downloads
Additional Files
Published
Issue
Section
Categories
How to Cite
Accepted 2015-06-11
Published 2015-06-23









