Long noncoding RNA RP11-70C1.3 confers chemoresistance of breast cancer cells through miR-6736-3p/NRP-1 axis
DOI:
https://doi.org/10.17305/bjbms.2021.5803Keywords:
Breast cancer, RP11-70C1.3, chemoresistance, NRP-1, miR-6736-3pAbstract
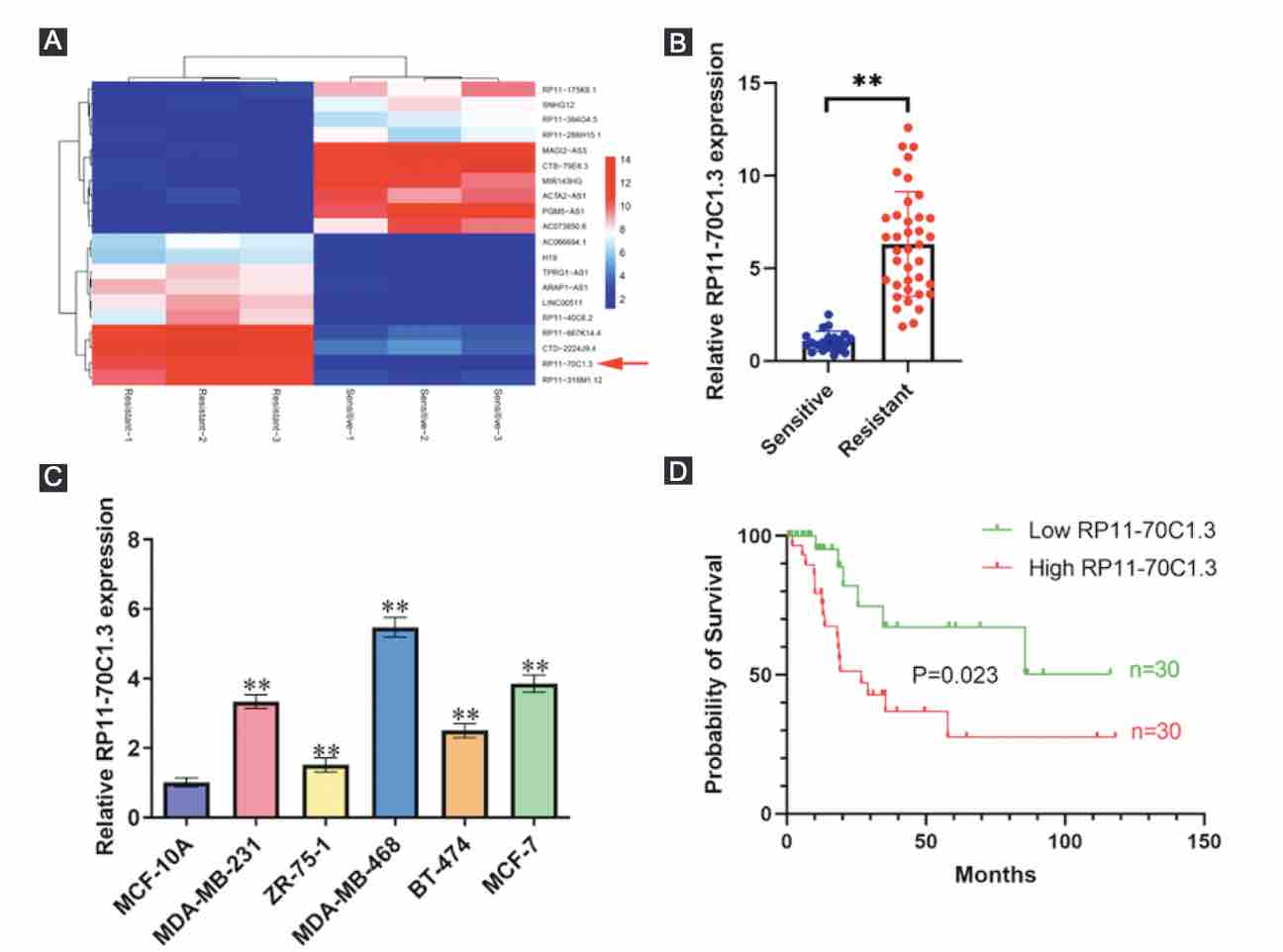
Chemoresistance remains a major obstacle for improving the clinical outcome of patients with breast cancer. Recently, long noncoding RNAs (lncRNAs) have been implicated in breast cancer chemoresistance. However, the function and underlying mechanism are still largely unknown. Using lncRNA microarray, we identified 122 upregulated and 475 downregulated lncRNAs that might be related to the breast cancer chemoresistance. Among them, RP11-70C1.3 was one of the most highly expressed lncRNAs. In breast cancer patients, high RP11-70C1.3 expression predicted poor prognosis. Knockdown of RP11-70C1.3 inhibited the multidrug resistance of breast cancer cells in vitro and in vivo. Further investigations revealed that RP11-70C1.3 functioned as a competing endogenous RNA (ceRNA) for miR-6736-3p to increase NRP-1 expression. Notably, the rescue experiments showed that both miR-6736-3p inhibitor and NRP-1 overexpression could partly reverse the suppressive influence of RP11-70C1.3 knockdown on breast cancer chemoresistance. In conclusion, our study indicated that lncRNA RP11-70C1.3 regulated NRP-1 expression by sponging miR-6736-3p to confer chemoresistance of breast cancer cells. RP11-70C1.3 might be a potential therapeutic target in enhancing the clinical efficacy of chemotherapy in breast cancer.
Citations
Downloads
Downloads
Additional Files
Published
Issue
Section
Categories
How to Cite
Accepted 2021-06-09
Published 2022-02-01









