Global DNA methylation and chondrogenesis of rat limb buds in a three-dimensional organ culture system
DOI:
https://doi.org/10.17305/bjbms.2021.6584Keywords:
Chondrogenesis, global DNA methylation, limb bud, development, rat, organ-culture, ex vivo, in vitro, serum-free, embryoAbstract
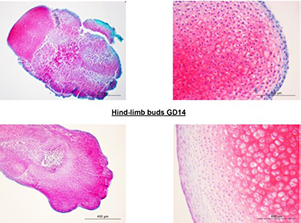
Although DNA methylation epigenetically regulates development, data on global DNA methylation during development of limb buds (LBs) are scarce. We aimed to investigate the global DNA methylation developmental dynamics in rat LBs cultivated in a serum-supplemented (SS) and in chemically defined serum- and protein-free (SF) three-dimensional organ culture. Fischer rat front- and hind-LBs at 13th and 14th gestation days (GD) were cultivated at the air-liquid interface in Eagle's Minimal Essential Medium (MEM) or MEM with 50% rat serum for 14 days, as SF and SS conditions, respectively. The methylation of repetitive DNA sequences (SINE rat ID elements) was assessed by pyrosequencing. Development was evaluated by light microscopy and extracellular matrix glycosaminoglycans staining by Safranin O. Upon isolation, weak Safranin O staining was present only in more developed GD14 front-LBs. Chondrogenesis proceeded well in all cultures towards day 14, except in the SF-cultivated GD13 hind-LBs, where Safranin O staining was almost absent on day 3. That was associated with a higher percentage of DNA methylation than in SF-cultivated GD13 front-LBs on day three. In SF-cultivated front-LBs, a significant methylation increase between the 3rd and 14th day was detected. In SS-cultivated GD13 front-LBs, methylation increased significantly on day three and then decreased. In older GD14 SS-cultivated LBs, there was no increase of DNA methylation, but they were significantly hypomethylated relative to the SS-cultivated GD13 at days 3 and 14. We confirmed that the global DNA methylation increase is associated with less developed limb organ primordia that strive towards differentiation in vitro, which is of importance for regenerative medicine strategies.
Citations
Downloads
Downloads
Additional Files
Published
License
Copyright (c) 2022 Vedrana Mužić Radović, Paula Bunoza, Tihana Marić, Marta Himelreich-Perić, Floriana Bulić-Jakuš, Marta Takahashi, Gordana Jurić-Lekić, Nino Sinčić, Davor Ježek, Ana Katušić-Bojanac

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2021-12-22
Published 2022-07-29









