Clinical and histopathological characteristics of COL4A3 c.2881+1G>A variant causing Alport spectrum disorders in Croatian population
DOI:
https://doi.org/10.17305/bjbms.2022.7567Keywords:
Alport syndrome, thin basement membrane nephropathy, proteinuria, collagen type IV, α3 chain of collagen IV, COL4A3 c.2881 1G>A variant, targeted next-generation sequencingAbstract
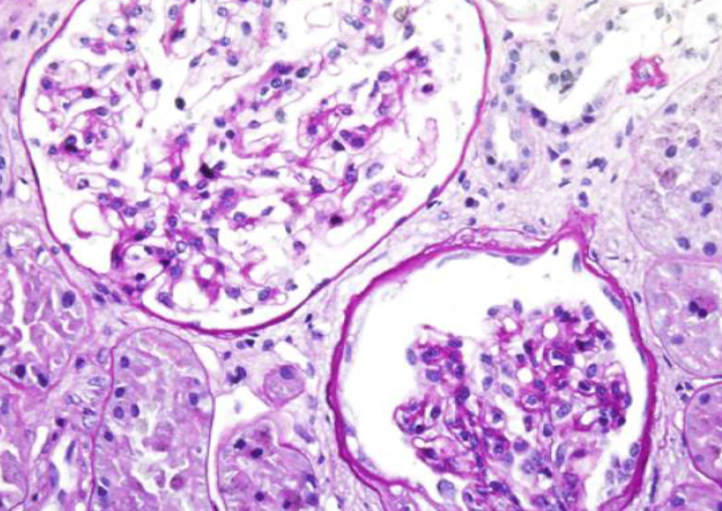
Alport syndrome (AS) and thin basement membrane nephropathy (TBMN) are part of the spectrum of kidney disorders caused by pathogenic variants in α3, α4, or α5 chains of the collagen type IV, the major structural component of the glomerular basement membrane (GBM). Using targeted next-generation sequencing (NGS), 34 AS/TBMN patients (58.8% male) from 12 unrelated families were found positive for heterozygous c.2881+1G>A variant of the COL4A3gene, that is considered disease-causing. All patients were from the continental or island part of Croatia. Clinical, laboratory, and histopathological data collected from the medical records were analyzed and compared to understand the clinical course and prognosis of the affected patients. At the time of biopsy or first clinical evaluation, the mean age was 31 years (median: 35 years; range: 1 – 72 years). Hematuria was present in 33 patients (97.1%) and 19 (55.9%) patients had proteinuria. There were 6 (17.6%) patients with hearing loss, 4 (11.8%) with ocular lesions, and 11 (32.4%) with hypertension. Twenty-three (67.6%) patients had proteinuria at follow-up, and 5 (14.7%) patients with the median age of 48 years (range: 27-55) progressed to kidney failure, started dialysis, or underwent kidney transplantation. Of the 13 patients who underwent kidney biopsy, 4 (30.8%) developed focal segmental glomerulosclerosis (FSGS), and 8 (66.7%) showed lamellation of the GBM, including all patients with FSGS. It is essential to conduct a detailed analysis of each collagen type IV genetic variant to optimize the prognosis and therapeutic approach for affected patients.
Citations
Downloads
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2022 Matija Horaček, Tamara Nikuševa Martić, Petar Šenjug, Marija Šenjug Perica, Maja Oroz, Sania Kuzmac, Dragan Klarić, Merica Glavina Durdov, Marijan Saraga, Danko Milošević, Danica Batinić, Marijana Ćorić, Frane Paić, Danica Galešić Ljubanović

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2022-06-28
Published 2023-01-06









