Promotive role of USP29-mediated deubiquitination in malignant proliferation of colorectal cancer cells via the KIAA1429/SOX8 axis
DOI:
https://doi.org/10.17305/bjbms.2022.7930Keywords:
Colorectal cancer, USP29, KIAA1429, SOX8, TPD52, malignant proliferationAbstract
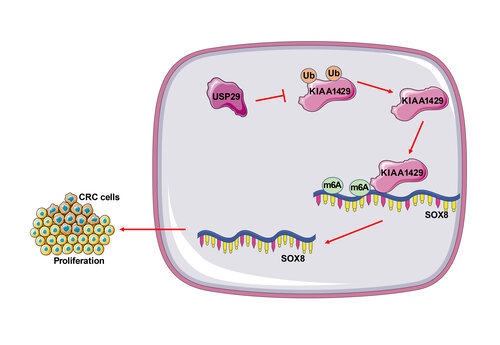
Colorectal cancer (CRC) is regarded as one of the most prevalent neoplasms worldwide, and ubiquitination and N6-methyladenosine (m6A) modification regulate the outgrowth of multiple cancers. This study attempted to explore the effect of ubiquitin-specific peptidases 29 (USP29) on the malignant proliferation of CRC cells via stabilizing Vir-like m6A methyltransferase associated (VIRMA/KIAA1429). First, upregulations of USP29, KIAA1429, and SRY-box transcription factor 8 (SOX8) were found in CRC tissues and cells through real-time quantitative polymerase chain reaction and Western blotting. After transfection of si-USP29, the proliferation of CRC cells was evaluated by the cell counting kit-8, colony formation and 5-ethynyl-2’-deoxyuridine assays, and we observed that depletion of USP29 inhibited the proliferation of CRC cells. Co-immunoprecipitation confirmed the binding of USP29 to KIAA1429. Mechanically, USP29 mediated deubiquitination to stabilize the protein levels of KIAA1429, and KIAA1429 promoted the stability of SOX8 mRNA through m6A modification. Moreover, overexpression of KIAA1429 or SOX8 reversed the inhibitory effects of USP29 depletion on CRC cell proliferation. Finally, the xenograft tumor model revealed the promotive role of USP29 in the proliferation of CRC cells in vivo. Altogether, USP29 facilitates the malignant proliferation of CRC cells via upregulating the KIAA1429/SOX8 axis.
Citations
Downloads
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2022 Juncai Li, Jianguo Yang, Zhenzhou Chen, Li Liu, Hao Wang, Qican Deng, Yajun Chen, Yong Que, Zhongxue Fu

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2022-10-12
Published 2023-05-01









