Artificial intelligence in renal pathology: Current status and future
DOI:
https://doi.org/10.17305/bjbms.2022.8318Keywords:
Artificial intelligence, deep learning, digital pathology, nephropathology, image analysis, convolutional neural networksAbstract
Renal biopsy pathology is an essential gold standard for the diagnosis of most kidney diseases. With the increase in the incidence rate of kidney diseases, the lack of renal pathologists, and an imbalance in their distribution, there is an urgent need for a new renal pathological diagnosis model. Advances in artificial intelligence (AI) along with the growing digitization of pathology slides for diagnosis are promising approach to meet the demand for more accurate detection, classification, and prediction of the outcome of renal pathology. AI has contributed substantially to a variety of clinical applications, including renal pathology. Deep learning, a subfield of AI that is highly flexible and supports automatic feature extraction, is increasingly being used in multiple areas of pathology. In this narrative review, we first provide a general description of AI methods, and then discuss the current and prospective applications of AI in the field of renal pathology. Both diagnostic and predictive prognostic applications are covered, emphasizing AI in renal pathology images, predictive models, and 3D in renal pathology. Finally, we outline the challenges associated with the implementation of AI platforms in renal pathology and provide our perspective on how these platforms might change in this field.
Citations
Downloads
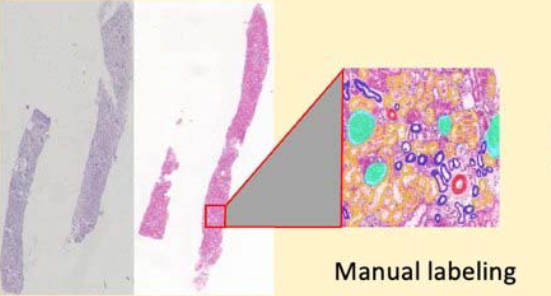
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2022 Chunyue Feng, Fei Liu

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2022-11-05
Published 2023-03-16









