Deciphering transcriptional dynamics of cardiac hypertrophy and failure in a chamber-specific manner
DOI:
https://doi.org/10.17305/bb.2023.8997Keywords:
Cardiac hypertrophy (CH), heart failure (HF), transcriptome, dynamic changes, pathogenesisAbstract
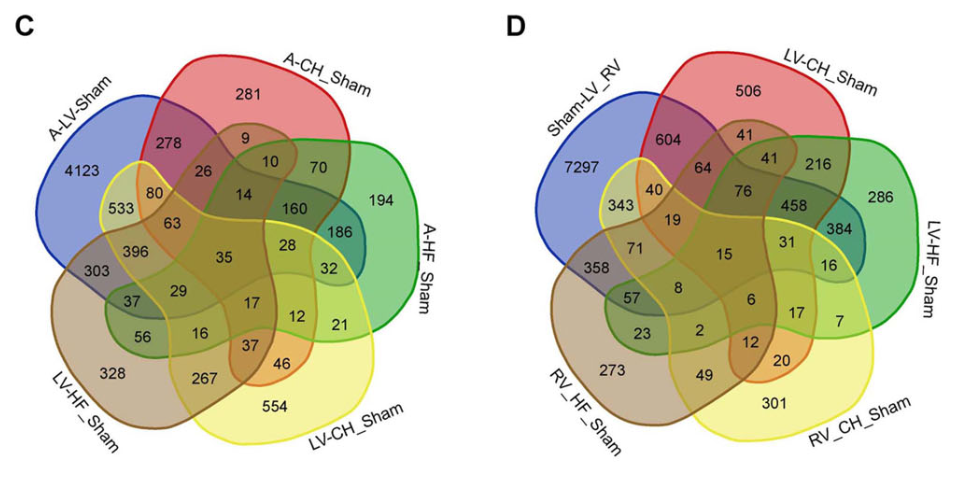
Pressure overload-induced pathological cardiac hypertrophy (CH) is a complexed and adaptive remodeling of the heart, predominantly involving an increase in cardiomyocyte size and thickening of ventricular walls. Over time, these changes can lead to heart failure (HF). However, the individual and shared biological mechanisms of both processes remain poorly understood. This study aimed to identify key genes and signaling pathways associated with CH and HF following aortic arch constriction (TAC) at four weeks and six weeks, respectively, and to investigate potential underlying molecular mechanisms in this dynamic transition from CH to HF at the whole cardiac transcriptome level. Initially, a total of 363, 482, and 264 differentially expressed genes (DEGs) for CH, and 317, 305, and 416 DEGs for HF were identified in the left atrium (LA), left ventricle (LV), and right ventricle (RV), respectively. These identified DEGs could serve as biomarkers for the two conditions in different heart chambers. Additionaly, two communal DEGs, elastin (ELN) and hemoglobin beta chain-beta S variant (HBB-BS), were found in all chambers, with 35 communal DEGs in the LA and LV and 15 communal DEGs in the LV and RV in both CH and HF. Functional enrichment analysis of these genes emphasized the crucial roles of the extracellular matrix and sarcolemma in CH and HF. Lastly, three groups of hub genes, including the lysyl oxidase (LOX) family, fibroblast growth factors (FGF) family, and NADH-ubiquinone oxidoreductase (NDUF) family, were determined to be essential genes of dynamic changes from CH to HF.
Citations
Downloads
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2023 Dan Zhang, Jianming Liu, Haiying Xiao, Jun Li, Ling Cao, Guang Li

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2023-06-09
Published 2023-11-03









