Comprehensive exosomal microRNA profile and construction of competing endogenous RNA network in autism spectrum disorder: A pilot study
DOI:
https://doi.org/10.17305/bb.2023.9552Keywords:
Autism spectrum disorder, exosome, miRNA, ceRNA, immune cell infiltrationAbstract
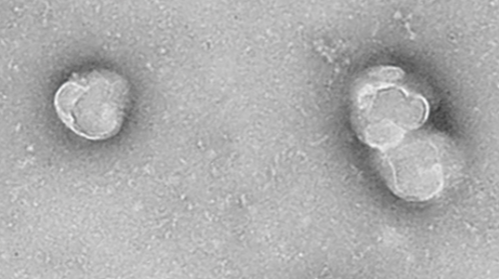
Exosomes have been demonstrated to exert momentous roles in autism spectrum disorder (ASD). However, few studies have reported a correlation between exosomal microRNAs (miRNAs) and ASD. To date, our understanding of crucial competing endogenous RNA (ceRNA) networks in ASD remains limited. Herein, the exosomal miRNA profile in the peripheral blood of children with ASD and healthy controls was investigated and the level of immune cell infiltration in ASD was evaluated to determine the distribution of immune cell subtypes. Exosomes were isolated from the peripheral blood of ten children with ASD and ten healthy controls, and further identified using transmission electron microscopy and western blot analysis. RNA sequencing was conducted to investigate exosomal miRNA profiles in patients with ASD. The mRNA and circular RNA (circRNA) expression profiles were acquired from the Gene Expression Omnibus (GEO) database. Differentially expressed mRNAs (DEmRNAs), miRNAs (DEmiRNAs), and circRNAs (DEcircRNAs) were identified and ceRNA regulatory networks were constructed. Furthermore, the immune cell infiltration levels in patients with ASD were evaluated. Exosomes were spherical, approximately 100 nm in size, and were confirmed via western blot analysis using exosome-associated markers CD9, CD63, and CD81. Thirty-five DEmRNAs, 63 DEmiRNAs, and 494 DEcircRNAs were identified in patients with ASD. CeRNA regulatory networks, including 6 DEmRNAs, 14 DEmiRNAs, and 86 DEcircRNAs, were established. Correlation analysis indicated that leucine-rich glioma inactivated protein 1 (LGI1) expression was significantly positively correlated with the content of CD8+ T cells. Our findings may be conducive to offering novel insights into this disease and providing further evidence of transcriptomic abnormalities in ASD.
Citations
Downloads
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2023 Sha Zhao, Yan Zhong, Fang Shen, Xinning Cheng, Xiaojuan Qing, Jiamin Liu

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2023-09-18
Published 2024-03-11









