Whole-exome sequencing in obstructive coronary artery disease identifies rare and novel variants in cardiac arrhythmia and pulmonary arterial hypertension–associated genes
DOI:
https://doi.org/10.17305/bb.2026.13200Keywords:
Whole exome sequencing, coronary artery disease, familial hypercholesterolemia, cardiac arrhythmias, pulmonary arterial hypertension, genetic variations, human phenotypeAbstract
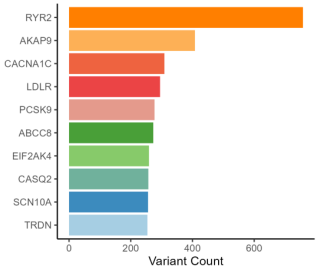
Coronary artery disease (CAD) represents a complex interplay of genetic, environmental, and lifestyle factors. In this study, we utilized whole-exome sequencing (WES) on 28 patients with obstructive CAD to identify rare variants that may influence clinical outcomes beyond conventional atherosclerotic risk. We examined 74 genes curated from the Genomics England PanelApp, focusing on familial hypercholesterolemia (FH), cardiac arrhythmias (CA), and pulmonary arterial hypertension (PAH), ultimately detecting 8,251 variants. After applying a stringent filtering process with a population maximum allele frequency (PopMax AF) threshold of <0.1%, we identified 68 candidate variants across 23 genes. The majority were associated with CA (47/68, 69%), followed by PAH (12/68, 18%) and FH (9/68, 13%). Notably, 30 variants (44%) were novel, and 18 were categorized as high-impact frameshift mutations. The highest burden of candidate variants was found in the sodium voltage-gated channel alpha subunit 10 (SCN10A), followed by the ryanodine receptor 2 (RYR2), mitochondrial seryl-tRNA synthetase 2 (SARS2), A-kinase anchoring protein 9 (AKAP9), and hyperpolarization-activated cyclic nucleotide-gated channel 4 (HCN4). Clinical evaluation revealed a pathogenic variant in the low-density lipoprotein receptor (LDLR) and likely pathogenic variants in sodium voltage-gated channel alpha subunit 5 (SCN5A) and potassium voltage-gated channel subfamily Q member 1 (KCNQ1); additionally, nine other variants were predicted to be deleterious, including five novel SCN10A variants. Functional annotation using Gene Ontology (GO) and Human Phenotype Ontology (HPO) highlighted mechanisms impacting cardiac structure, electrical conduction, and lipid homeostasis.
Citations
Downloads
References
World Health Organization. Cardiovascular diseases (CVDs) [Internet]. Geneva: World Health Organization; 2025 Jul 31 [cited 2026 Jan 14]. Available from:
https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds)
Libby P, Buring JE, Badimon L, et al. Atherosclerosis. Nat Rev Dis Primers. 2019;5(1):56.
https://doi.org/10.1038/s41572-019-0106-z
Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;370(25):2418–2425.
https://doi.org/10.1056/NEJMra1312543
Khera AV, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–1224.
https://doi.org/10.1038/s41588-018-0183-z
Natarajan P, Young R, Stitziel NO, et al. PCSK9 loss-of-function variants and risk of coronary heart disease: 5,716 cases and 20,675 controls. JAMA Cardiol. 2017;2(2):155–162.
Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Internet]. 2010 [cited 2026 Jan 14]. Available from:
http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Poplin R, Ruano-Rubio V, DePristo MA, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. 2017:201178.
https://doi.org/10.1101/201178
McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17(1):122.
https://doi.org/10.1186/s13059-016-0974-4
Liu X, Li C, Mou C, et al. dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020;12:103.
https://doi.org/10.1186/s13073-020-00803-9
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894.
https://doi.org/10.1093/nar/gky1016
Köhler S, Gargano M, Matentzoglu N, et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 2021;49(D1):D1207–D1217.
https://doi.org/10.1093/nar/gkaa1043
Online Mendelian Inheritance in Man, OMIM®. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD). 2023.
Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–D985.
https://doi.org/10.1093/nar/gkt1113
UniProt Consortium. The universal protein resource (UniProt). Nucleic Acids Res. 2008;36(Database issue):D190–D195.
https://doi.org/10.1093/nar/gkm895
Fairley S, Lowy-Gallego E, Perry E, Flicek P. The International Genome Sample Resource (IGSR) collection of open human genomic variation resources. Nucleic Acids Res. 2020;48(D1):D941–D947.
https://doi.org/10.1093/nar/gkz836
Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443.
Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874.
https://doi.org/10.1101/gr.176601
Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249.
https://doi.org/10.1038/nmeth0410-248
Rogers MF, Shihab HA, Mort M, et al. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics. 2018;34(3):511–513.
https://doi.org/10.1093/bioinformatics/btx536
Schwarz J, Cooper D, Schuelke M, et al. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2024;11:361–362.
https://doi.org/10.1038/nmeth.2890
Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118.
https://doi.org/10.1093/nar/gkr407
Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31(16):2745–2747.
https://doi.org/10.1093/bioinformatics/btv195
Hu J, Ng PC. SIFT Indel: predictions for the functional effects of amino acid insertions/deletions in proteins. PLoS One. 2013;8(10):e77940.
https://doi.org/10.1371/journal.pone.0077940
Martin AR, Williams E, Foulger RE, et al. PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat Genet. 2019;51(11):1560–1565.
https://doi.org/10.1038/s41588-019-0528-2
Kolberg L, Raudvere U, Kuzmin I, et al. g:Profiler—interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023;51(W1):W207–W212.
https://doi.org/10.1093/nar/gkad347
Stark B, Johnson C, Roth G. Global prevalence of coronary artery disease: an update from the Global Burden of Disease Study. J Am Coll Cardiol. 2024;83(13 Suppl):2320.
https://doi.org/10.1016/S0735-1097(24)04310-9
Bottardi A, Prado GFA, Lunardi M, et al. Clinical updates in coronary artery disease: a comprehensive review. J Clin Med. 2024;13(16):4600.
https://doi.org/10.3390/jcm13164600
Nakazato R, Arsanjani R, Achenbach S, et al. Age-related risk of major adverse cardiac event risk and coronary artery disease extent and severity by coronary CT angiography: results from 15,187 patients from the International Multisite CONFIRM Study. Eur Heart J Cardiovasc Imaging. 2014;15:586–594.
https://doi.org/10.1093/ehjci/jet132
Nordström A, Bergman J, Björk S, et al. A multiple risk factor program is associated with decreased risk of cardiovascular disease in 70-year-olds: a cohort study from Sweden. PLoS Med. 2020;17:e1003135.
https://doi.org/10.1371/journal.pmed.1003135
Vaura F, Palmu J, Aittokallio J, et al. Genetic, molecular, and cellular determinants of sex-specific cardiovascular traits. Circ Res. 2022;130(4):611–631.
https://doi.org/10.1161/CIRCRESAHA.121.319891
Hsu SP, Lee WS. Effects of female sex hormones on the development of atherosclerosis. Chin J Physiol. 2020;63(6):256–262.
https://doi.org/10.4103/CJP.CJP_69_20
Bots SH, Peters SAE, Woodward M. Sex differences in coronary heart disease and stroke mortality: a global assessment of the effect of ageing between 1980 and 2010. BMJ Glob Health. 2017;2(2):e000298.
https://doi.org/10.1136/bmjgh-2017-000298
Canto JG, Rogers WJ, Goldberg RJ, et al. Association of age and sex with myocardial infarction symptom presentation and in-hospital mortality. JAMA. 2012;307(8):813–822.
https://doi.org/10.1001/jama.2012.199
Sakkers TR, Mokry M, Civelek M, et al. Sex differences in the genetic and molecular mechanisms of coronary artery disease. Atherosclerosis. 2023;384:117279.
https://doi.org/10.1016/j.atherosclerosis.2023.117279
Yusuf S, Hawken S, Ounpuu S, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364:937–952.
https://doi.org/10.1016/S0140-6736(04)17018-9
Weber T, Lang I, Zweiker R, et al. Hypertension and coronary artery disease: epidemiology, physiology, effects of treatment, and recommendations: a joint scientific statement from the Austrian Society of Cardiology and the Austrian Society of Hypertension. Wien Klin Wochenschr. 2016;128(13–14):467–479.
https://doi.org/10.1007/s00508-016-0998-5
Zang J, Liang J, Zhuang X, et al. Intensive blood pressure treatment in coronary artery disease: implications from the Systolic Blood Pressure Intervention Trial (SPRINT). J Hum Hypertens. 2022;36(1):86–94.
https://doi.org/10.1038/s41371-021-00494-8
Lewington S, Clarke R, Qizilbash N, et al. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360:1903–1913.
https://doi.org/10.1016/S0140-6736(02)11911-8
Ference BA, Ginsberg HN, Graham I, et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease: evidence from genetic, epidemiologic, and clinical studies: a consensus statement from the European Atherosclerosis Society Consensus Panel. Eur Heart J. 2017;38:2459–2472.
https://doi.org/10.1093/eurheartj/ehx144
Kathiresan S, Manning AK, Demissie S, et al. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S17.
https://doi.org/10.1186/1471-2350-8-S1-S17
Genest JJ Jr, Martin-Munley SS, McNamara JR, et al. Familial lipoprotein disorders in patients with premature coronary artery disease. Circulation. 1992;85:2025–2033.
https://doi.org/10.1161/01.CIR.85.6.2025
Rämö JT, Ripatti P, Tabassum R, et al. Coronary artery disease risk and lipidomic profiles are similar in hyperlipidemias with family history and population-ascertained hyperlipidemias. J Am Heart Assoc. 2019;8(13):e012415.
https://doi.org/10.1161/JAHA.119.012415
Atique SM, Shadbolt B, Marley P, Farshid A. Association between body mass index and age of presentation with symptomatic coronary artery disease. Clin Cardiol. 2016;39:653–657.
https://doi.org/10.1002/clc.22576
Ades PA. Cardiac rehabilitation and secondary prevention of coronary heart disease. N Engl J Med. 2001;345:892–902.
https://doi.org/10.1056/NEJMra001529
Yoon JW, Jung CH, Kim MK, et al. Influence of the definition of “metabolically healthy obesity” on the progression of coronary artery calcification. PLoS One. 2017;12:e0178741.
https://doi.org/10.1371/journal.pone.0178741
Manoharan MP, Raja R, Jamil A, et al. Obesity and coronary artery disease: an updated systematic review 2022. Cureus. 2022;14(9):e29480.
https://doi.org/10.7759/cureus.29480
American Diabetes Association Professional Practice Committee. Cardiovascular disease and risk management: Standards of Care in Diabetes—2024. Diabetes Care. 2024;47(Suppl 1):S179–S218.
Fowler MJ. Microvascular and macrovascular complications of diabetes. Clin Diabetes. 2008;26:77–82.
https://doi.org/10.2337/diaclin.26.2.77
Ashraf FUN, Ghouri K, Someshwar F, et al. Insulin resistance and coronary artery disease: untangling the web of endocrine-cardiac connections. Cureus. 2023;15(12):e51066.
https://doi.org/10.7759/cureus.51066
Joseph JJ, Deedwania P, Acharya T, et al. Comprehensive management of cardiovascular risk factors for adults with type 2 diabetes: a scientific statement from the American Heart Association. Circulation. 2022;145(9):e722–e759.
https://doi.org/10.1161/CIR.0000000000001040
Khunti K, Kosiborod M, Ray KK. Legacy benefits of blood glucose, blood pressure and lipid control in individuals with diabetes and cardiovascular disease: time to overcome multifactorial therapeutic inertia? Diabetes Obes Metab. 2018;20:1337–1341.
https://doi.org/10.1111/dom.13243
Di Lenarda F, Balestrucci A, Terzi R, et al. Coronary artery disease, family history, and screening perspectives: an up-to-date review. J Clin Med. 2024;13(19):5833.
https://doi.org/10.3390/jcm13195833
Nielsen M, Andersson C, Gerds TA, et al. Familial clustering of myocardial infarction in first-degree relatives: a nationwide study. Eur Heart J. 2013;34(16):1198–1203.
https://doi.org/10.1093/eurheartj/ehs475
Vrints C, Andreotti F, Koskinas KC, et al; ESC Scientific Document Group. 2024 ESC Guidelines for the management of chronic coronary syndromes. Eur Heart J. 2024;45(36):3415–3537.
Iyer KR, Clarke SL, Guarischi-Sousa R, et al. Unveiling the genetic landscape of coronary artery disease through common and rare structural variants. J Am Heart Assoc. 2025;14(4):e036499.
Marshall CR, Chowdhury S, Taft RJ, et al. Best practices for the analytical validation of clinical whole-genome sequencing intended for the diagnosis of germline disease. NPJ Genom Med. 2020;5:47.
https://doi.org/10.1038/s41525-020-00154-9
Scrocco C, Conte G, Behr ER. The application of genomics in the arrhythmia clinic. e-J Cardiol Pract. 2022;22(15).
Scrocco C, Bezzina CR, Ackerman MJ, Behr ER. Genetics and genomics of arrhythmic risk: current and future strategies to prevent sudden cardiac death. Nat Rev Cardiol. 2021;18:774–784.
https://doi.org/10.1038/s41569-021-00555-y
Sturm AC, Knowles JW, Gidding SS, et al; Convened by the Familial Hypercholesterolemia Foundation. Clinical genetic testing for familial hypercholesterolemia: JACC Scientific Expert Panel. J Am Coll Cardiol. 2018;72(6):662–680.
https://doi.org/10.1016/j.jacc.2018.05.044
Fahed AC, Wang M, Patel AP, et al. Association of the interaction between familial hypercholesterolemia variants and adherence to a healthy lifestyle with risk of coronary artery disease. JAMA Netw Open. 2022;5(3):e222687.
https://doi.org/10.1001/jamanetworkopen.2022.2687
Gallo G, Savoia C. Hypertension and heart failure: from pathophysiology to treatment. Int J Mol Sci. 2024;25(12):6661.
https://doi.org/10.3390/ijms25126661
Kraja AT, Hunt SC, Rao DC, et al. Genetics of hypertension and cardiovascular disease and their interconnected pathways: lessons from large studies. Curr Hypertens Rep. 2011;13(1):46–54.
https://doi.org/10.1007/s11906-010-0174-7
Ginsberg HN, Fisher EA. The ever-expanding role of degradation in the regulation of apolipoprotein B metabolism. J Lipid Res. 2009;50(Suppl):S162–S166.
https://doi.org/10.1194/jlr.R800090-JLR200
Singh K, Prabhakaran D. Apolipoprotein B—an ideal biomarker for atherosclerosis? Indian Heart J. 2024;76(Suppl 1):S121–S129.
https://doi.org/10.1016/j.ihj.2023.12.001
Sharma R, Mahajan M, Singh B, et al. Role of the APOB gene polymorphism (c.12669G>A, p.Gln4154Lys) in coronary artery disease in the Indian Punjabi population. Balkan J Med Genet. 2011;14(2):35–40.
https://doi.org/10.2478/v10034-011-0045-9
Gallegos-Arreola MP, Valdez Y, Zúñiga-Corona M, et al. Association between the Xba I polymorphism of APOB gene and plasma lipid level in Mexican patients with coronary artery disease. Asia Pac J Clin Nutr. 2012;21(2):312–318.
Shen H, Damcott CM, Rampersaud E, et al. Familial defective apolipoprotein B-100 and increased low-density lipoprotein cholesterol and coronary artery calcification in the Old Order Amish. Arch Intern Med. 2010;170(20):1850–1855.
https://doi.org/10.1001/archinternmed.2010.384
Han C, Huang J, Waxman SG. Sodium channel Nav1.8: emerging links to human disease. Neurology. 2016;86(5):473–483.
https://doi.org/10.1212/WNL.0000000000002333
Behr ER, Savio-Galimberti E, Barc J, et al; UK10K Consortium. Role of common and rare variants in SCN10A: results from the Brugada syndrome QRS locus gene discovery collaborative study. Cardiovasc Res. 2015;106(3):520–529.
https://doi.org/10.1093/cvr/cvv042
Huang Y, Chen XM, Barajas-Martinez H, et al. Common variants in SCN10A gene associated with Brugada syndrome. Hum Mol Genet. 2021;31(2):157–165.
https://doi.org/10.1093/hmg/ddab217
Suryavanshi SV, Jadhav SM, McConnell BK. Polymorphisms/mutations in A-kinase anchoring proteins (AKAPs): role in the cardiovascular system. J Cardiovasc Dev Dis. 2018;5(1):7.
https://doi.org/10.3390/jcdd5010007
Ercu M, Klussmann E. Roles of A-kinase anchoring proteins and phosphodiesterases in the cardiovascular system. J Cardiovasc Dev Dis. 2018;5(1):14.
https://doi.org/10.3390/jcdd5010014
Allegue C, Coll M, Mates J, et al. Genetic analysis of arrhythmogenic diseases in the era of NGS: the complexity of clinical decision-making in Brugada syndrome. PLoS One. 2015;10(7):e0133037.
https://doi.org/10.1371/journal.pone.0133037
Forleo C, D’Erchia AM, Sorrentino S, et al. Targeted next-generation sequencing detects novel gene-phenotype associations and expands the mutational spectrum in cardiomyopathies. PLoS One. 2017;12(7):e0181842.
https://doi.org/10.1371/journal.pone.0181842
Guo L, Wu D, Zhu W. Reappraisal of ANK2 variants in cardiovascular diseases: uncovering mechanisms and future directions. Rev Cardiovasc Med. 2025;26(1):26013.
https://doi.org/10.31083/RCM26013
Koenig SN, Mohler PJ. The evolving role of ankyrin-B in cardiovascular disease. Heart Rhythm. 2017;14(12):1884–1889.
https://doi.org/10.1016/j.hrthm.2017.07.032
Steinberg C, Roston TM, van der Werf C, et al. RYR2-ryanodinopathies: from calcium overload to calcium deficiency. Europace. 2023;25(6):euad156.
https://doi.org/10.1093/europace/euad156
Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291.
https://doi.org/10.1038/nature19057
Vouloagkas I, Agbariah A, Zegkos T, et al. The many faces of SCN5A pathogenic variants: from channelopathy to cardiomyopathy. Heart Fail Rev. 2025;30(1):247–256.
https://doi.org/10.1007/s10741-024-10459-x
Morgat C, Fressart V, Porretta AP, et al. Genetic characterization of KCNQ1 variants improves risk stratification in type 1 long QT syndrome patients. Europace. 2024;26(6):euae136.
Downloads
Published
Issue
Section
Categories
License
Copyright (c) 2026 Mohammad Fahad Ullah, Rashid Mir, Jamsheed Javid, Imadeldin Elfaki, Faisal H. Altemani, Jameel Barnawi, Naseh A. Algehainy, Mohammed M. Jalal, Malik A. Altayar, Salem Owaid Albalawi, Syed Khalid Mustafa, Aadil Yousif, Eram Husain, Faris J. Tayeb, Faisel M. AbuDuhier

This work is licensed under a Creative Commons Attribution 4.0 International License.









