Genetic risk of alcohol-related liver cirrhosis: Associations of PNPLA3, TM6SF2, and a two-variant polygenic risk score
DOI:
https://doi.org/10.17305/bb.2025.13261Keywords:
Alcohol-related liver cirrhosis, ALC, genetic variants, PNPLA3, TM6SF2, polygenic risk score, PRSAbstract
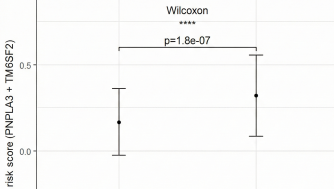
A minority of individuals who consume excessive alcohol develop cirrhosis. Variants in the patatin-like phospholipase domain-containing protein 3 gene (PNPLA3) and the transmembrane 6 superfamily member 2 gene (TM6SF2) have been previously identified as associated with alcohol-related cirrhosis (ALC). This study aimed to examine the variants of PNPLA3 and TM6SF2 and to develop and assess a polygenic risk score (PRS) for ALC. We enrolled 118 patients diagnosed with ALC and 131 control subjects, who were either abstainers or low-level alcohol consumers without evidence of liver disease. Genotyping of risk variants was performed using PCR-RFLP methodology. PRS, based on independent allelic effect size estimates from genotyped genetic loci, were computed and compared across groups. The development of ALC was significantly associated with CG and GG genotypes of PNPLA3 (CG: OR: 1.82; 95% CI: 1.05-3.17; p=0.033; GG: OR: 7.64; 95% CI: 3.06-19.07; p<0.001) and the CT genotype of TM6SF2 (OR: 2.43; 95% CI: 1.27-4.63; p=0.007), controlling for age and sex. Patients with cirrhosis exhibited a significantly higher mean PRS compared to controls (0.32 vs. 0.167, p = 1.8e-07). The odds ratios (ORs) and 95% confidence intervals for the group with the highest PRS score compared to the reference group were 6.707; 95% CI: 3.313-13.581, p<0.001. In our ALC patient cohort, the PNPLA3 rs738409 and TM6SF2 rs58542926 variants were associated with an increased risk of ALC development. Moreover, the PRS derived from these two variants effectively identified the genetic components linked to cirrhosis within the study population.
Citations
Downloads
References
Teli, M.R., Day, C.P., Burt, A.D., Bennett, M.K., James, O.F.W., Determinants of progression to cirrhosis or fibrosis in pure alcoholic fatty liver. Lancet, 1995. 346(8981): p. 987–990. https://doi.org/10.1016/S0140-6736(95)91685-7
Ginès, P., Krag, A., Abraldes, J.G., Solà, E., Fabrellas, N., Kamath, P.S., Liver cirrhosis. Lancet, 2021. 398(10308): p. 1359–1376. https://doi.org/10.1016/S0140-6736(21)01374-X
Devarbhavi, H., Singh, S.P., Khillan, V., Reddy, K.R., Chawla, Y.K., Bhattacharya, P., et al., Global burden of liver disease: 2023 update. J Hepatol, 2023. 79(2): p. 516–537. https://doi.org/10.1016/j.jhep.2023.03.017
Simpson, R.F., Shipley, M.J., Griffin, S.J., Brunner, E.J., Cairns, B.J., Beral, V., et al., Alcohol drinking patterns and liver cirrhosis risk: analysis of the prospective UK Million Women Study. Lancet Public Health, 2019. 4(1): p. e41–e48. https://doi.org/10.1016/S2468-2667(18)30230-5
Wood, A.M., Kaptoge, S., Butterworth, A.S., Willeit, P., Warnakula, S., Bolton, T., et al., Risk thresholds for alcohol consumption: combined analysis of individual-participant data for 599,912 current drinkers in 83 prospective studies. Lancet, 2018. 391(10129): p. 1513–1523. https://doi.org/10.1016/S0140-6736(18)30134-X
Huang, D.Q., El-Serag, H.B., Loomba, R., Global epidemiology of alcohol-associated cirrhosis and HCC: trends, projections and risk factors. Nat Rev Gastroenterol Hepatol, 2023. 20(1): p. 37–49. https://doi.org/10.1038/s41575-022-00688-6
Hrubec, Z., Omenn, G.S., Evidence of genetic predisposition to alcoholic cirrhosis and psychosis: twin concordances for alcoholism and its biological end points by zygosity among male veterans. Alcohol Clin Exp Res, 1981. 5(2): p. 207–215. https://doi.org/10.1111/j.1530-0277.1981.tb04890.x
Reed, T., Dick, D.M., Slutske, W.S., Heath, A.C., Bierut, L.J., Saccone, N.L., et al., Genetic predisposition to organ-specific endpoints of alcoholism. Alcohol Clin Exp Res, 1996. 20(9): p. 1528–1533. https://doi.org/10.1111/j.1530-0277.1996.tb01695.x
Konkwo, C., Chowdhury, S., Vilarinho, S., Genetics of liver disease in adults. Hepatol Commun, 2024. 8(4). https://doi.org/10.1097/HC9.0000000000000408
Stickel, F., Datz, C., Hampe, J., Bataller, R., Rehm, J., Parés, A., et al., Pathophysiology and management of alcoholic liver disease: update 2016. Gut Liver, 2017. 11(2): p. 173–188. https://doi.org/10.5009/gnl16477
Buch, S., Stickel, F., Trépo, E., Way, M., Herrmann, A., Nischalke, H.D., et al., Variants in PCSK7, PNPLA3 and TM6SF2 are risk factors for the development of cirrhosis in hereditary haemochromatosis. Aliment Pharmacol Ther, 2021. 53(7): p. 830–843. https://doi.org/10.1111/apt.16252
Buch, S., Stickel, F., Trépo, E., Way, M., Herrmann, A., Nischalke, H.D., et al., A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet, 2015. 47(12): p. 1443–1448. https://doi.org/10.1038/ng.3417
Falleti, E., Bitetto, D., Fabris, C., Cmet, S., Cillo, U., Cussigh, A., et al., PNPLA3 rs738409 and TM6SF2 rs58542926 variants increase the risk of hepatocellular carcinoma in alcoholic cirrhosis. Dig Liver Dis, 2016. 48(1): p. 69–75. https://doi.org/10.1016/j.dld.2015.09.009
Kozlitina, J., Smagris, E., Stender, S., Nordestgaard, B.G., Zhou, H., Tybjærg-Hansen, A., et al., Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet, 2014. 46(4): p. 352–356. https://doi.org/10.1038/ng.2901
Huang, Y., He, S., Li, J., Li, X., Chao, Y., Li, J., et al., A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc Natl Acad Sci U S A, 2010. 107(17): p. 7892–7897. https://doi.org/10.1073/pnas.1003585107
Galvanin, C., Dongiovanni, P., Meroni, M., Baselli, G., Romeo, S., Valenti, L., et al., PNPLA3 in alcohol-related liver disease. Liver Int, 2025. 45(1): p. e16211. https://doi.org/10.1111/liv.16211
Romeo, S., Kozlitina, J., Xing, C., Pertsemlidis, A., Cox, D., Pennacchio, L.A., et al., Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet, 2008. 40(12): p. 1461–1465. https://doi.org/10.1038/ng.257
Tian, C., Stokowy, T., Li, J., Zeng, Q., Jin, L., Li, X., et al., Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet, 2010. 42(1): p. 21–23. https://doi.org/10.1038/ng.488
Dongiovanni, P., Valenti, L., Anstee, Q.M., Petta, S., Maglio, C., Pipitone, R., et al., PNPLA3 I148M polymorphism and progressive liver disease. World J Gastroenterol, 2013. 19(41): p. 6969–6978. https://doi.org/10.3748/wjg.v19.i41.6969
Trépo, E., Nahon, P., Bontempi, G., Valenti, L., Falleti, E., Colombo, M., et al., Common polymorphism in the PNPLA3/adiponutrin gene confers higher risk of cirrhosis and liver damage in alcoholic liver disease. J Hepatol, 2011. 55(4): p. 906–912. https://doi.org/10.1016/j.jhep.2011.01.028
Liu, J., Ginsberg, H.N., Reyes-Soffer, G., Basic and translational evidence supporting the role of TM6SF2 in VLDL metabolism. Curr Opin Lipidol, 2024. 35(3): p. 157–161. https://doi.org/10.1097/MOL.0000000000000930
Meroni, M., Dongiovanni, P., Baselli, G., Valenti, L., Romeo, S., Anstee, Q.M., et al., Genetic and epigenetic modifiers of alcoholic liver disease. Int J Mol Sci, 2018. 19(12): p. 3857. https://doi.org/10.3390/ijms19123857
Shihana, F., Galvanin, C., Dongiovanni, P., Baselli, G., Valenti, L., Romeo, S., et al., Investigating the role of lipid genes in liver disease using fatty liver models of alcohol and high fat in zebrafish (Danio rerio). Liver Int, 2023. 43(11): p. 2455–2468. https://doi.org/10.1111/liv.15716
Julien, J., Mansuy-Aubert, V., Thabut, D., Ratziu, V., Cadranel, J.F., Trépo, E., et al., Effect of increased alcohol consumption during COVID-19 pandemic on alcohol-associated liver disease: a modeling study. Hepatology, 2022. 75(6): p. 1480–1490. https://doi.org/10.1002/hep.32272
Kann, A.E., Ruhl, C.E., Mozaffarian, D., Schmidt, H., Rehm, J., Whitfield, J.B., et al., Motivation to reduce drinking and engagement in alcohol misuse treatment in alcohol-related liver disease: a national health survey. Am J Gastroenterol, 2022. 117(6): p. 918–922. https://doi.org/10.14309/ajg.0000000000001616
World Health Organization, International guide for monitoring alcohol consumption and related harm. Geneva: World Health Organization, 2000.
Enssle, J., Weylandt, K.H., Secure and optimized detection of PNPLA3 rs738409 genotype by an improved PCR-restriction fragment length polymorphism method. Biotechniques, 2021. 70(6): p. 345–349. https://doi.org/10.2144/btn-2020-0163
Urzua, A., González, A., Morales, A., Villanueva, J., Poniachik, J., Rojas, A., et al., TM6SF2 rs58542926 polymorphism is not associated with risk of steatosis or fibrosis in Chilean patients with chronic hepatitis C. Hepat Mon, 2017. 17(4): p. e12307. https://doi.org/10.5812/hepatmon.44365
Sinnott-Armstrong, N., Tanigawa, Y., Amar, D., Mars, N., Benner, C., Aguirre, M., et al., Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat Genet, 2021. 53: p. 185–194. https://doi.org/10.1038/s41588-020-00757-z
Moon, S.Y., Park, J., Kim, H., Lee, J., Song, Y., Choi, S., et al., Alcohol consumption and the risk of liver disease: a nationwide, population-based study. Front Med (Lausanne), 2023. 10: p. 1290266. https://doi.org/10.3389/fmed.2023.1290266
Corrao, G., Bagnardi, V., Zambon, A., Arico, S., Poikolainen, K., La Vecchia, C., et al., Meta-analysis of alcohol intake in relation to risk of liver cirrhosis. Alcohol Alcohol, 1998. 33(4): p. 381–392. https://doi.org/10.1093/oxfordjournals.alcalc.a008408
Roerecke, M., Rehm, J., Patra, J., Popova, S., Zatonski, W., Przybylski, R., et al., Alcohol consumption and risk of liver cirrhosis: a systematic review and meta-analysis. Am J Gastroenterol, 2019. 114(10): p. 1574–1586. https://doi.org/10.14309/ajg.0000000000000340
Stickel, F., Hampe, J., Genetic determinants of alcoholic liver disease. Gut, 2012. 61(1): p. 150–159. https://doi.org/10.1136/gutjnl-2011-301239
Salameh, H., Meroni, M., Dongiovanni, P., Valenti, L., Romeo, S., Baselli, G., et al., PNPLA3 gene polymorphism is associated with predisposition to and severity of alcoholic liver disease. Am J Gastroenterol, 2015. 110(6): p. 846–856. https://doi.org/10.1038/ajg.2015.137
Rashu, E.B., Meroni, M., Galvanin, C., Valenti, L., Romeo, S., Dongiovanni, P., et al., Use of PNPLA3, TM6SF2, and HSD17B13 for detection of fibrosis in MASLD in the general population. Clin Res Hepatol Gastroenterol, 2024. 48(7): p. 102389. https://doi.org/10.1016/j.clinre.2024.102389
Stickel, F., Trépo, E., Meroni, M., Dongiovanni, P., Baselli, G., Valenti, L., et al., Genetic variants in PNPLA3 and TM6SF2 predispose to the development of hepatocellular carcinoma in individuals with alcohol-related cirrhosis. Am J Gastroenterol, 2018. 113(10): p. 1475–1483. https://doi.org/10.1038/s41395-018-0041-8
Trépo, E., Nahon, P., Bontempi, G., Valenti, L., Falleti, E., Colombo, M., et al., PNPLA3 gene in liver diseases. J Hepatol, 2016. 65(2): p. 399–412. https://doi.org/10.1016/j.jhep.2016.03.011
Kumari, M., Xu, X., Chai, J., Brindley, D.N., Wang, Y., Li, Z., et al., Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase. Cell Metab, 2012. 15(5): p. 691–702. https://doi.org/10.1016/j.cmet.2012.04.008
Bainrauch, A., Galvanin, C., Trépo, E., Dongiovanni, P., Meroni, M., Valenti, L., et al., NOTCH3 rs1043996 polymorphism is associated with the occurrence of alcoholic liver cirrhosis independently of PNPLA3 and TM6SF2 polymorphisms. J Clin Med, 2021. 10(19): p. 4621. https://doi.org/10.3390/jcm10194621
Falleti, E., Bitetto, D., Cmet, S., Fabris, C., Cillo, U., Cussigh, A., et al., PNPLA3 rs738409C/G polymorphism in cirrhosis: relationship with the aetiology of liver disease and hepatocellular carcinoma occurrence. Liver Int, 2011. 31(8): p. 1137–1143. https://doi.org/10.1111/j.1478-3231.2011.02534.x
Nischalke, H.D., Berg, T., Luda, C., Zimmermann, T., Odenthal, M., Müller, T., et al., The PNPLA3 rs738409 148M/M genotype is a risk factor for liver cancer in alcoholic cirrhosis but shows no or weak association in hepatitis C cirrhosis. PLoS One, 2011. 6(11): p. e27087. https://doi.org/10.1371/journal.pone.0027087
Rosendahl, J., Becker, M., Tönjes, A., Sowa, J.P., Schulze, F., Teumer, A., et al., A common variant of PNPLA3 (p.I148M) is not associated with alcoholic chronic pancreatitis. PLoS One, 2012. 7(1): p. e29433. https://doi.org/10.1371/journal.pone.0029433
Fairfield, C.J., O'Reilly, M., Simon, T.G., Harrison, S.A., Younossi, Z., Anstee, Q.M., et al., Genome-wide association study of NAFLD using electronic health records. Hepatol Commun, 2022. 6(2): p. 297–308. https://doi.org/10.1002/hep4.1805
Genome Aggregation Database (gnomAD), Available online: https://gnomad.broadinstitute.org (accessed on 10th March 2025).
Karczewski, K.J., Francioli, L.C., Tiao, G., Cummings, B.B., Alföldi, J., Wang, Q., et al., The mutational constraint spectrum quantified from variation in 141,456 humans. Nature, 2020. 581(7809): p. 434–443. https://doi.org/10.1038/s41586-020-2308-7
Whitfield, J.B., Heath, A.C., Madden, P.A., Montgomery, G.W., Martin, N.G., Statham, D.J., et al., A genetic risk score and diabetes predict development of alcohol-related cirrhosis in drinkers. J Hepatol, 2022. 76(2): p. 275–282. https://doi.org/10.1016/j.jhep.2021.10.005
Ghouse, J., Kühn, F., Nissen, P.H., Andersson, C., Sørensen, K.M., Hildebrandt, P., et al., Integrative common and rare variant analyses provide insights into the genetic architecture of liver cirrhosis. Nat Genet, 2024. 56(5): p. 827–837. https://doi.org/10.1038/s41588-024-01720-y
Schwantes-An, T.H., Wu, Y., McGowan, C.R., Palmer, C.N., Whitfield, J.B., Daly, A.K., et al., A polygenic risk score for alcohol-associated cirrhosis among heavy drinkers with European ancestry. Hepatol Commun, 2024. 8(6): p. e0431.
Chambers, J.C., Zhang, W., Sehmi, J., Li, X., Wass, M.N., Van der Harst, P., et al., Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet, 2011. 43(11): p. 1131–1138. https://doi.org/10.1038/ng.970
Viitasalo, A., Lindi, V., Schwab, U., Tompuri, T., Laaksonen, D.E., Kautiainen, H., et al., Associations of I148M variant in PNPLA3 gene with plasma ALT levels during 2-year follow-up in normal weight and overweight children: the PANIC Study. Pediatr Obes, 2015. 10(2): p. 84–90. https://doi.org/10.1111/ijpo.234
Dai, G., Zhu, Y., Li, H., Zhang, Y., Li, Z., Wang, Y., et al., Association between PNPLA3 rs738409 polymorphism and nonalcoholic fatty liver disease (NAFLD) susceptibility and severity: a meta-analysis. Medicine (Baltimore), 2019. 98(7): p. e14324. https://doi.org/10.1097/MD.0000000000014324
Procopet, B., Berzigotti, A., Diagnosis of cirrhosis and portal hypertension: imaging, non-invasive markers of fibrosis and liver biopsy. Gastroenterol Rep (Oxf), 2017. 5(2): p. 79–89. https://doi.org/10.1093/gastro/gox012
Tevik, K., Haugland, S., Landmark-Høyvik, H., Krokstad, S., Holmen, T.L., Holmen, J., et al., A systematic review of self-report measures used in epidemiological studies to assess alcohol consumption among older adults. PLoS One, 2021. 16(12): p. e0261292. https://doi.org/10.1371/journal.pone.0261292
Downloads
Published
Issue
Section
Categories
License
Copyright (c) 2025 Branka Nesic, Marina Jelovac, Teodora Karan-Djurasevic, Dusica Vrinic Kalem, Petar Svorcan, Branka Zukic, Ivana Grubisa

This work is licensed under a Creative Commons Attribution 4.0 International License.









