Epidemiology and mechanism of drug resistance of Mycoplasma pneumoniae in Beijing, China: A multicenter study
DOI:
https://doi.org/10.17305/bjbms.2019.4053Keywords:
Mycoplasma pneumoniae, epidemiology, drug resistance, infectionAbstract
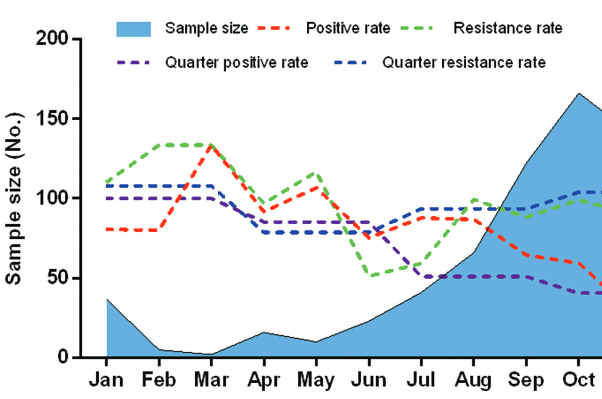
Mycoplasma pneumoniae (M. pneumoniae) is one of the most common causes of community-acquired respiratory tract infections (RTIs). We aimed to investigate the prevalence of M. pneumoniae infection, antibiotic resistance and genetic diversity of M. pneumoniae isolates across multiple centers in Beijing, China. P1 protein was detected by Nested PCR to analyze the occurrence of M. pneumoniae in pediatric patients with RTI. M. pneumoniae isolates were cultured and analyzed by Nested-PCR to determine their genotypes. Broth microdilution method was used to determine the minimum inhibitory concentration (MIC) of antibiotics. Out of 822 children with RTI admitted to 11 hospitals in Beijing, 341 (41.48%) were positive for M. pneumoniae by Nested PCR and 236 (69.21%) samples had mutations in 23S rRNA domain V. The highest proportion of M. pneumoniae positive samples was observed in school-age children (118/190; 62.11%) and in pediatric patients with pneumonia (220/389; 56.56%). Out of 341 M. pneumoniae positive samples, 99 (12.04%) isolates were successfully cultured and the MIC values were determined for 65 M. pneumoniae strains. Out of these, 57 (87.69%) strains were resistant to macrolides, and all 65 strains were sensitive to tetracyclines or quinolones. M. pneumoniae P1 type I and P1 type II strains were found in 57/65 (87.69%) and 8/65 (12.31%) of cultured isolates, respectively. Overall, we demonstrated a high prevalence of M. pneumoniae infection and high macrolide resistance of M. pneumoniae strains in Beijing. School-age children were more susceptible to M. pneumoniae, particularly the children with pneumonia. Thus, establishment of a systematic surveillance program to fully understand the epidemiology of M. pneumoniae is critical for the standardized use of antibiotics in China.
Citations
Downloads
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2019 Bosnian Journal of Basic Medical Sciences

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2019-01-07
Published 2019-08-20









