The prognostic value of MKL1 in predicting breast cancer immune infiltrates and chemosensitivity
DOI:
https://doi.org/10.17305/bjbms.2021.6306Keywords:
MKL1, breast cancer, immune cell infiltration, chemosensitivityAbstract
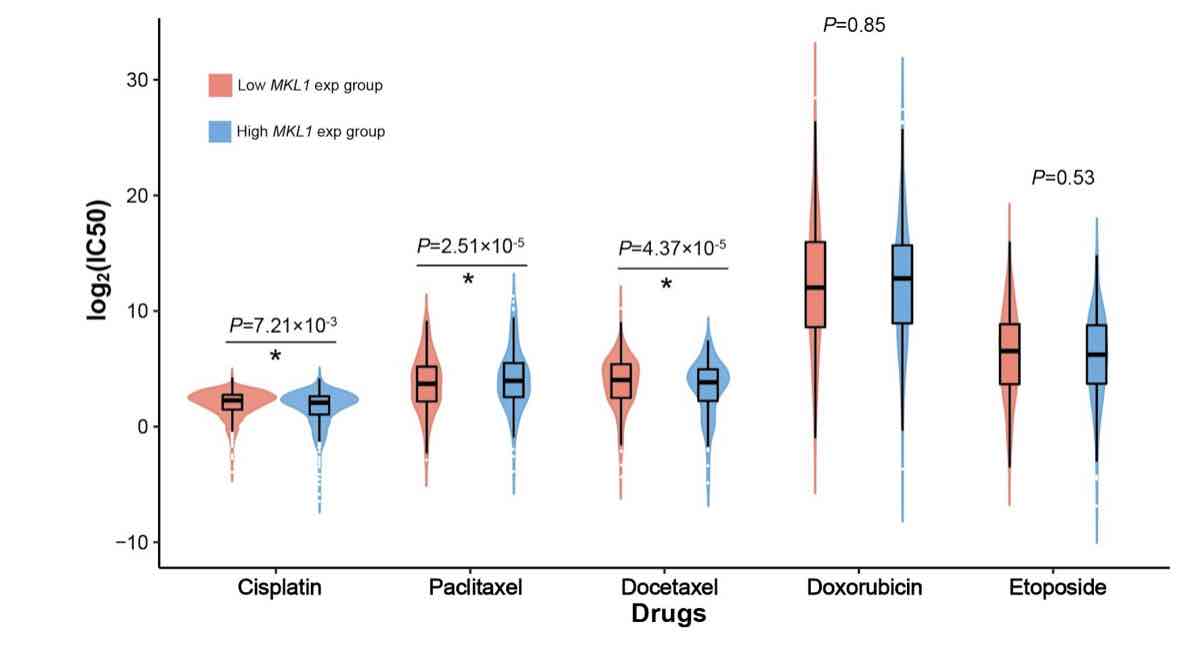
Megakaryocytic leukemia 1 (MKL1) acts as a transcription factor in the regulation of the immune system and is associated with cancer biology. However, its function in the infiltrating immune cells in breast cancer has not been explored. Our study aimed to analyze the expression of MKL1 in The Cancer Genome Atlas (TCGA) breast cancer dataset. The aim of this study was to evaluate the correlations between MKL1 expression, infiltrating immune cells, and immune control genes. Enriched signaling pathways and drug sensitivity analyses were also performed. Our results indicate that high MKL1 expression could predict better survival in breast cancer patients. MKL1 expression was associated with the expression and function of different immune cells, including T cells, B cells, natural killer (NK) cells, macrophages, neutrophils and dendritic cells (DCs). The chromatin modifying enzymes, cellular senescence, epigenetic regulation of gene expression, estrogen-dependent gene expression, and chromosome maintenance were differentially enriched in MKL1 low expression phenotype. Patients in the high MKL1 expression group showed sensitivity to paclitaxel, while those in the low expression group showed potential sensitivity for cisplatin and docetaxel. In conclusion, MKL1 might act as a potential biomarker of prognostic value for immune infiltration and drug sensitivity in breast cancer.
Citations
Downloads
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2021 Yijia Hua, Mengzhu Yang

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2021-11-07
Published 2022-06-01









