Whole-exome sequencing reveals rare genetic variations in ovarian granulosa cell tumor
DOI:
https://doi.org/10.17305/bjbms.2021.6789Keywords:
Whole-exome sequencing, ovarian granulosa cell tumor, single-nucleotide polymorphism, indels, ovarian cancerAbstract
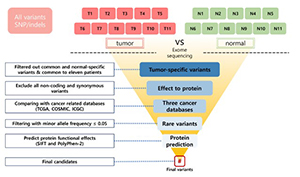
Ovarian granulosa cell tumor (OGCT) is a rare ovarian tumor that accounts for about 2-5% of all ovarian tumors. Despite the low grade of ovarian tumors, high and late recurrences are common in OGCT patients. Even though this tumor usually occurs in adult women with high estrogen levels, the cause of OGCT is still unknown. To screen genetic variants associated with OGCT, we collected normal and matched-tumor formalin-fixed paraffin-embedded (FFPE) from 11 OGCT patients and performed whole-exome sequencing (WES) using Illumina NovaSeq 6000. A total of 1,067,219 single nucleotide polymorphisms (SNPs) and 162,155 insertions/deletions (indels) were identified from 11 pairs of samples. Of these, we identified 44 tumor-specific SNPs in 22 genes and four tumor-specific indels in one gene that were common to 11 patients. We used three cancer databases (TCGA, COSMIC, and ICGC) to investigate genes associated with ovarian cancers. Nine genes (SEC22B, FEZ2, ANKRD36B, GYPA, MUC3A, PRSS3, NUTM2A, OR8U1, and KRTAP10-6) associated with ovarian cancers were found in all three databases. In addition, we identified seven rare variants with MAF ≤ 0.05 in two genes (PRSS3 and MUC3A). Of seven rare variants, five variants in MUC3A are potentially pathogenic. Furthermore, we conducted gene enrichment analysis of tumor-specific 417 genes in SNPs and 106 genes in indels using cytoscape and metascape. In GO analysis, these genes were highly enriched in “selective autophagy”, and “regulation of anoikis”. Taken together, we suggest that MUC3A is implicated in OGCT development, and MUC3A could be used as a potential biomarker for OGCT diagnosis.
Citations
Downloads
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2022 Seungyeon Kim, Songmi Kim, Seyoung Mun, Yongsik Kwak, Kwang-Sun Suh, Song-Yi Choi, Kyudong Han

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2021-12-27
Published 2022-06-01









