S-palmitoylation-related genes in Crohn's disease: Bioinformatic identification and validation
DOI:
https://doi.org/10.17305/bb.2025.13221Keywords:
Crohn’s disease, S-palmitoylation, ZDHHC23, IFITM1, immune infiltration, machine learningAbstract
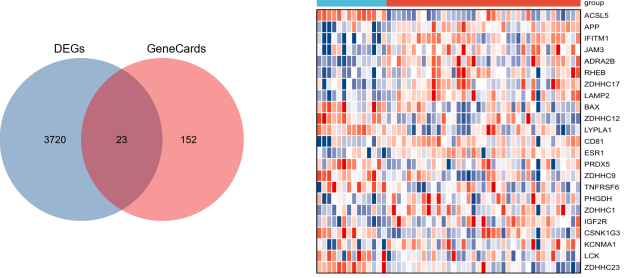
Crohn's disease (CD) is a complex chronic inflammatory bowel disorder characterized by the absence of reliable biomarkers and effective targeted treatments. Recent evidence has suggested a role for S-palmitoylation, a reversible post-translational modification, in immune regulation and intestinal inflammation. However, a systematic, gene-centric investigation explicitly linking S-palmitoylation to the pathogenesis and diagnosis of CD has not been conducted. To address this gap, our study employs a comprehensive bioinformatic analysis to identify and validate key genes associated with both CD and S-palmitoylation, assessing their potential as diagnostic biomarkers and therapeutic targets. Utilizing data from the Gene Expression Omnibus (GEO, GSE83448) and GeneCards, we identified 23 S-palmitoylation-associated differentially expressed genes (SP-DEGs) in CD. Functional enrichment analysis indicated their significant roles in cysteine-specific S-palmitoylation and immunometabolic regulation. We applied machine learning algorithms, including least absolute shrinkage and selection operator (LASSO) regression and support vector machine–recursive feature elimination (SVM-RFE), to select nine hub genes. Validation in two independent cohorts (GSE16879 and GSE59071) and ROC analysis confirmed ZDHHC23 and IFITM1 as biomarkers with high diagnostic value. These genes also exhibited correlations with immune infiltration patterns, as determined by cell-type identification by estimating relative subsets of RNA transcripts (CIBERSORT), MCPcounter, and QuanTIseq. In vitro experiments corroborated consistent changes in mRNA and protein expression for both ZDHHC23 and IFITM1, reinforcing their involvement in CD. This study offers systematic insights into the functional roles of S-palmitoylation-related genes in CD, providing a novel theoretical foundation for the development of diagnostic and targeted therapeutic strategies.
Citations
Downloads
References
Sazonovs A, Stevens CR, Venkataraman GR, Yuan K, Avila B, Abreu MT, et al. Large-scale sequencing identifies multiple genes and rare variants associated with Crohn's disease susceptibility. Nat Genet 2022;54(9):1275–83. https://doi.org/10.1038/s41588-022-01156-2.
Cao S, Colonna M, Deepak P. Pathogenesis of Perianal Fistulising Crohn's Disease: Current Knowledge, Gaps in Understanding, and Future Research Directions. J Crohns Colitis 2023;17(6):1010–22. https://doi.org/10.1093/ecco-jcc/jjad008.
Gisbert JP, Chaparro M. Anti-TNF Agents and New Biological Agents (Vedolizumab and Ustekinumab) in the Prevention and Treatment of Postoperative Recurrence After Surgery in Crohn's Disease. Drugs 2023;83(13):1179–205. https://doi.org/10.1007/s40265-023-01916-2.
Zheng M, Han R, Yuan Y, Xing Y, Zhang W, Sun Z, et al. The role of Akkermansia muciniphila in inflammatory bowel disease: Current knowledge and perspectives. Front Immunol 2022;13:1089600. https://doi.org/10.3389/fimmu.2022.1089600.
Ding NS, Hart A, De Cruz P. Systematic review: predicting and optimising response to anti-TNF therapy in Crohn's disease – algorithm for practical management. Aliment Pharmacol Ther 2016;43(1):30–51. https://doi.org/10.1111/apt.13445.
Foppa C, Rizkala T, Repici A, Hassan C, Spinelli A. Microbiota and IBD: Current knowledge and future perspectives. Dig Liver Dis 2024;56(6):911–22. https://doi.org/10.1016/j.dld.2023.11.015.
Mesquita FS, Abrami L, Linder ME, Bamji SX, Dickinson BC, van der Goot FG. Mechanisms and functions of protein S-acylation. Nat Rev Mol Cell Biol 2024;25(6):488–509. https://doi.org/10.1038/s41580-024-00700-8.
Main A, Fuller W. Protein S-Palmitoylation: advances and challenges in studying a therapeutically important lipid modification. FEBS J 2022;289(4):861–82. https://doi.org/10.1111/febs.15781.
Zhang MM, Hang HC. Protein S-palmitoylation in cellular differentiation. Biochem Soc Trans 2017;45(1):275–85. https://doi.org/10.1042/bst20160236.
Chen R, Tang X, Wang Y, Wang B, Mao F. Protein palmitoylation: an emerging regulator of inflammatory signaling and diseases. Front Immunol 2025;16:1652741. https://doi.org/10.3389/fimmu.2025.1652741.
Das T, Yount JS, Hang HC. Protein S-palmitoylation in immunity. Open Biol 2021;11(3):200411. https://doi.org/10.1098/rsob.200411.
Li P, Gong X, Yuan L, Mu L, Zheng Q, Xiao H, Wang H. Palmitoylation in apoptosis. J Cell Physiol 2023;238(8):1641–50. https://doi.org/10.1002/jcp.31047.
Zhang Y, Qin Z, Sun W, Chu F, Zhou F. Function of Protein S-Palmitoylation in Immunity and Immune-Related Diseases. Front Immunol 2021;12:661202. https://doi.org/10.3389/fimmu.2021.661202.
Pei S, Piao HL. Exploring Protein S-Palmitoylation: Mechanisms, Detection, and Strategies for Inhibitor Discovery. ACS Chem Biol 2024;19(9):1868–82. https://doi.org/10.1021/acschembio.4c00110.
De I, Sadhukhan S. Emerging Roles of DHHC-mediated Protein S-palmitoylation in Physiological and Pathophysiological Context. Eur J Cell Biol 2018;97(5):319–38. https://doi.org/10.1016/j.ejcb.2018.03.005.
Cheng WX, Ren Y, Lu MM, Xu LL, Gao JG, Chen D, et al. Palmitoylation in Crohn's disease: Current status and future directions. World J Gastroenterol 2021;27(48):8201–15. https://doi.org/10.3748/wjg.v27.i48.8201.
Li H, Yuan Q, Wang S, Yu T, Qi X. Role of S-palmitoylation in digestive system diseases. Cell Death Discov 2025;11(1):331. https://doi.org/10.1038/s41420-025-02629-z.
Zheng S, Que X, Wang S, Zhou Q, Xing X, Chen L, et al. ZDHHC5-mediated NLRP3 palmitoylation promotes NLRP3-NEK7 interaction and inflammasome activation. Mol Cell 2023;83(24):4570–85.e7. https://doi.org/10.1016/j.molcel.2023.11.015.
Meng C, Dai X, Sun L, Huang D, Xu X, Cheng Y, Zhang W. ZDHHC21-driven S-palmitoylation of Themis regulates the function of T cells and maintains homeostatic balance. Cell Commun Signal 2025;23(1):401. https://doi.org/10.1186/s12964-025-02396-5.
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 2013;41:D991–5. https://doi.org/10.1093/nar/gks1193.
Liu S, Wang Z, Zhu R, Wang F, Cheng Y, Liu Y. Three Differential Expression Analysis Methods for RNA Sequencing: limma, EdgeR, DESeq2. J Vis Exp 2021;(175):62528. https://doi.org/10.3791/62528.
Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010;2010:baq020. https://doi.org/10.1093/database/baq020.
Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 2022;50(W1):W216–W221. https://doi.org/10.1093/nar/gkac194.
Tang L, Chen J, Wu Z, Wang L, Lai Y, Chen Z, et al. FUNDC1 predicts Poor Prognosis and promotes Progression and Chemoresistance in Endometrial Carcinoma. J Cancer 2024;15(20):6490–6504. https://doi.org/10.7150/jca.96877.
Ali H, Shahzad M, Sarfraz S, Sewell KB, Alqalyoobi S, Mohan BP. Application and impact of Lasso regression in gastroenterology: A systematic review. Indian J Gastroenterol 2023;42(6):780–90. https://doi.org/10.1007/s12664-023-01426-9.
Wang L, Wu P, Liu Y, Patel DC, Leonard TB, Zhao H. Clustering-aided prediction of outcomes in patients with idiopathic pulmonary fibrosis. Respir Res 2024;25(1):383. https://doi.org/10.1186/s12931-024-03015-6.
Sanz H, Valim C, Vegas E, Oller JM, Reverter F. SVM-RFE: selection and visualization of the most relevant features through non-linear kernels. BMC Bioinformatics 2018;19(1):432. https://doi.org/10.1186/s12859-018-2451-4.
Lu H, Lan S, Yuan X, Li P. Functional Enrichment, Drug Prediction, and Molecular Docking to Identify Fibroblast-Related Biomarkers for Gastric Cancer via High-Dimensional Weighted Gene Co-Expression Network Analysis. Endocr Metab Immune Disord Drug Targets 2025. https://doi.org/10.2174/0118715303424481250929070902.
Wang Z, Zhang Z. Biomarkers associated with cell-in-cell structure in kidney renal clear cell carcinoma based on transcriptome sequencing. PeerJ 2025;13:e19246. https://doi.org/10.7717/peerj.19246.
Yu G, Li F, Qin Y, Bo X, Wu Y, Wang S. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 2010;26(7):976–8. https://doi.org/10.1093/bioinformatics/btq064.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12(5):453–7. https://doi.org/10.1038/nmeth.3337.
Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020;577(7791):549–55. https://doi.org/10.1038/s41586-019-1922-8.
Finotello F, Mayer C, Plattner C, Laschober G, Rieder D, Hackl H, et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med 2019;11(1):34. https://doi.org/10.1186/s13073-019-0638-6.
Bruckner RS, Spalinger MR, Barnhoorn MC, Feakins R, Fuerst A, Jehle EC, et al. Contribution of CD3+CD8- and CD3+CD8+ T Cells to TNF-alpha Overexpression in Crohn Disease-Associated Perianal Fistulas and Induction of Epithelial-Mesenchymal Transition in HT-29 Cells. Inflamm Bowel Dis 2021;27(4):538–49. https://doi.org/10.1093/ibd/izaa240.
Pan X, Zhu Q, Pan LL, Sun J. Macrophage immunometabolism in inflammatory bowel diseases: From pathogenesis to therapy. Pharmacol Ther 2022;238:108176. https://doi.org/10.1016/j.pharmthera.2022.108176.
Adolph TE, Meyer M, Schwarzler J, Mayr L, Grabherr F, Tilg H. The metabolic nature of inflammatory bowel diseases. Nat Rev Gastroenterol Hepatol 2022;19(12):753–67. https://doi.org/10.1038/s41575-022-00658-y.
Aramsangtienchai P, Spiegelman NA, Cao J, Lin H. S-Palmitoylation of Junctional Adhesion Molecule C Regulates Its Tight Junction Localization and Cell Migration. J Biol Chem 2017;292(13):5325–34. https://doi.org/10.1074/jbc.m116.730523.
Oda Y, Sugawara T, Fukata Y, Izumi Y, Otani T, Higashi T, et al. The extracellular domain of angulin-1 and palmitoylation of its cytoplasmic region are required for angulin-1 assembly at tricellular contacts. J Biol Chem 2020;295(13):4289–302. https://doi.org/10.1074/jbc.ra119.010491.
Zingler P, Sarchen V, Glatter T, Caning L, Saggau C, Kathayat RS, et al. Palmitoylation is required for TNF-R1 signaling. Cell Commun Signal 2019;17(1):90. https://doi.org/10.1186/s12964-019-0405-8.
Feng Z, Meng F, Huo F, Zhu Y, Qin Y, Gui Y, et al. Inhibition of ferroptosis rescues M2 macrophages and alleviates arthritis by suppressing the HMGB1/TLR4/STAT3 axis in M1 macrophages. Redox Biol 2024;75:103255. https://doi.org/10.1016/j.redox.2024.103255.
Fan X, Zhang S, Sun S, Bi W, Li S, Wang W, et al. GFAP palmitoylation mediated by ZDHHC23 in spinal astrocytes contributes to the development of neuropathic pain. Reg Anesth Pain Med 2024;49(11):821–30. https://doi.org/10.1136/rapm-2023-104980.
Dai T, Zhao Z, Zhu T, Fei C, Nie L, Chen J. The anti-inflammatory role of zDHHC23 through the promotion of macrophage M2 polarization and macrophage necroptosis in large yellow croaker (Larimichthys crocea). Front Immunol 2024;15:1401626. https://doi.org/10.3389/fimmu.2024.1401626.
Barbi J, Pardoll D, Pan F. Treg functional stability and its responsiveness to the microenvironment. Immunol Rev 2014;259(1):115–39. https://doi.org/10.1111/imr.12172.
Wang K, Fu W. Transcriptional regulation of Treg homeostasis and functional specification. Cell Mol Life Sci 2020;77(21):4269–87. https://doi.org/10.1007/s00018-020-03534-7.
Tekguc M, Wing JB, Osaki M, Long J, Sakaguchi S. Treg-expressed CTLA-4 depletes CD80/CD86 by trogocytosis, releasing free PD-L1 on antigen-presenting cells. Proc Natl Acad Sci U S A 2021;118(30):e2023739118. https://doi.org/10.1073/pnas.2023739118.
Mitchell DA, Hamel LD, Reddy KD, Farh L, Rettew LM, Sanchez PR, Deschenes RJ. Mutations in the X-linked intellectual disability gene, zDHHC9, alter autopalmitoylation activity by distinct mechanisms. J Biol Chem 2014;289(26):18582–92. https://doi.org/10.1074/jbc.m114.567420.
Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res 2006;47(6):1118–27. https://doi.org/10.1194/jlr.r600007-jlr200.
Poggi M, Kara I, Brunel JM, Landrier JF, Govers R, Bonardo B, et al. Palmitoylation of TNF alpha is involved in the regulation of TNF receptor 1 signalling. Biochim Biophys Acta 2013;1833(3):602–12. https://doi.org/10.1016/j.bbamcr.2012.11.009.
Souza RF, Caetano MAF, Magalhaes HIR, Castelucci P. Study of tumor necrosis factor receptor in the inflammatory bowel disease. World J Gastroenterol 2023;29(18):2733–46. https://doi.org/10.3748/wjg.v29.i18.2733.
Waschke KA, Villani AC, Vermeire S, Dufresne L, Chen TC, Bitton A, et al. Tumor necrosis factor receptor gene polymorphisms in Crohn's disease: association with clinical phenotypes. Am J Gastroenterol 2005;100(5):1126–33. https://doi.org/10.1111/j.1572-0241.2005.40534.x.
Han J, Wu M, Liu Z. Dysregulation in IFN-gamma signaling and response: the barricade to tumor immunotherapy. Front Immunol 2023;14:1190333. https://doi.org/10.3389/fimmu.2023.1190333.
Song L, Wang D, Abbas G, Li M, Cui M, Wang J, et al. The main protease of SARS-CoV-2 cleaves histone deacetylases and DCP1A, attenuating the immune defense of the interferon-stimulated genes. J Biol Chem 2023;299(3):102990. https://doi.org/10.1016/j.jbc.2023.102990.
Nguyen HM, Gaikwad S, Oladejo M, Agrawal MY, Srivastava SK, Wood LM. Interferon stimulated gene 15 (ISG15) in cancer: An update. Cancer Lett 2023;556:216080. https://doi.org/10.1016/j.canlet.2023.216080.
Shi X, Chen S, Liu M, Fan Y, Wen X, Wang J, et al. The unconventional role of ABHD17A in increasing the S-palmitoylation and antiviral activity of IFITM1 by downregulating ABHD16A. Biomolecules 2025;15(7):992. https://doi.org/10.3390/biom15070992.
Wang Y, Shen N, Yang Y, Xia Y, Zhang W, Lu Y, et al. ZDHHC5-mediated S-palmitoylation of FAK promotes its membrane localization and epithelial-mesenchymal transition in glioma. Cell Commun Signal 2024;22(1):46. https://doi.org/10.1186/s12964-023-01366-z.
Zhou B, Hao Q, Liang Y, Kong E. Protein palmitoylation in cancer: molecular functions and therapeutic potential. Mol Oncol 2023;17(1):3–26. https://doi.org/10.1002/1878-0261.13308.
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2025 Yuyan Zhou, Yuxuan Zhao

This work is licensed under a Creative Commons Attribution 4.0 International License.









