Sepsis toxicity network reconstruction—Dynamic signaling and multi-organ injury: A review
DOI:
https://doi.org/10.17305/bb.2025.12931Keywords:
Sepsis, systemic toxicity, network biology, multiple organ dysfunction, dynamic evolution of signaling pathwaysAbstract
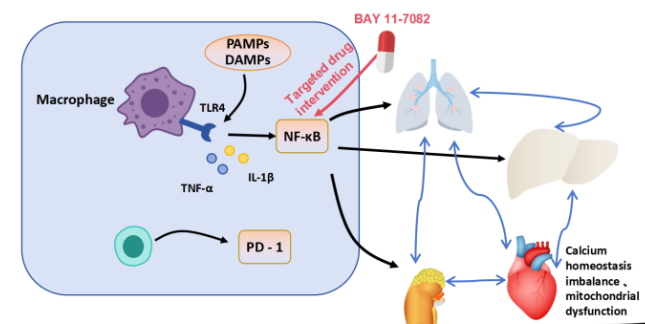
Sepsis is a complex systemic disease in which systemic toxicity—arising from inflammation–immune dysregulation, oxidative stress, programmed cell death (apoptosis, pyroptosis, ferroptosis), and metabolic reprogramming—drives multi-organ injury. The aim of this review was to synthesize how signaling pathways evolve within and between key organs (lungs, liver, kidneys, heart) and to evaluate whether multi-omics integration and network modeling can identify critical toxic nodes and predict disease progression. We conducted a narrative review of English-language mechanistic studies published between 2015 and 2025 in PubMed, Web of Science, and Scopus, supplemented by bibliography screening, while excluding case reports, conference abstracts, and non-mechanistic work. The evidence depicts a high-dimensional systemic network that remodels over time, with early pro-inflammatory modules transitioning toward immunosuppression and organ-specific injury patterns, while inter-organ propagation is mediated by damage-associated molecular patterns (DAMPs), exosomes, and metabolites. Oxidative stress and mitochondrial dysfunction, via reactive oxygen species (ROS), couple to pyroptosis and ferroptosis to reinforce toxicity loops, and computational approaches such as dynamic Bayesian networks (DBN) and graph neural networks (GNN) delineate regulatory hubs and support forecasting. Therapeutic progress has concentrated on nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), the NOD-, leucine-rich repeat and pyrin domain–containing protein 3 (NLRP3) inflammasome, and glutathione peroxidase 4 (GPX4), alongside artificial intelligence (AI)–assisted personalized toxicity maps and dynamic early-warning systems, though challenges remain in specificity, safety, and resistance. In conclusion, sepsis can be conceived as a temporally staged systemic toxicity network, and when combined with multi-omics, DBN/GNN modeling, and AI-enabled decision support, this framework offers a path toward individualized, mechanism-based care, while requiring rigorous validation to ensure clinical durability.
Citations
Downloads
Downloads
Published
Issue
Section
Categories
License
Copyright (c) 2025 Shuai Liu, Qun Liang

This work is licensed under a Creative Commons Attribution 4.0 International License.









