Pre-analytical storage effects on ALU- and LINE1-derived cell-free DNA biomarkers in whole blood and plasma
DOI:
https://doi.org/10.17305/bb.2026.13409Keywords:
Cell-free DNA, Arthrobacter luteus repeats, long interspersed nuclear elements 1, biomarkers, pre-analytical factorsAbstract
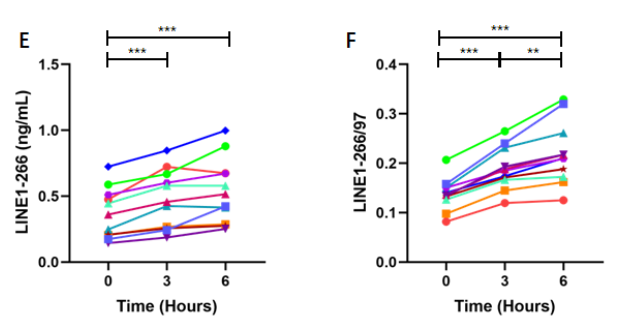
Cell-free DNA (cfDNA) biomarkers derived from Arthrobacter luteus (ALU) repeats and long interspersed nuclear elements 1 (LINE1) — including ALU-115, ALU-247, LINE1-97, and LINE1-266 concentrations, as well as the integrity ratios ALU-247/115 and LINE1-266/97 — are commonly utilized to assess cfDNA quantity and integrity. This study examined the impact of delayed blood processing and prolonged plasma storage on these biomarkers using quantitative polymerase chain reaction. Blood samples were collected from twelve healthy individuals (6 males; mean age, 65.8 ± 4.69 years) into dipotassium ethylenediaminetetraacetic acid tubes. Plasma cfDNA was extracted after various storage durations and temperatures, with aliquots from immediately processed blood subsequently stored at -80°C for different time intervals. Except for LINE1-97, most biomarkers showed significantly higher levels in plasma isolated from whole blood stored at room temperature compared to plasma processed immediately. Storage at 4°C resulted in fragment-specific effects: ALU-247/115 levels remained stable at 3 hours but decreased at 6 hours, while LINE1-266/97 levels increased at both time points. For plasma stored at -80°C, ALU-derived biomarkers remained stable for up to 12 months; however, LINE1-97 levels significantly declined, accompanied by a corresponding increase in LINE1-266/97 as early as one month after freezing. These findings indicate that both storage duration and temperature significantly impact the measured levels of ALU- and LINE1-derived cfDNA biomarkers. Consequently, standardization of pre-analytical handling of blood and plasma is crucial for studies evaluating cfDNA quantity and integrity.
Citations
Downloads
References
Han DSC, Lo YMD. The nexus of cfDNA and nuclease biology. Trends in Genetics. 2021;37:758–70.
https://doi.org/10.1016/j.tig.2021.04.005
Kamel AM, Teama S, Fawzy A, El Deftar M. Plasma DNA integrity index as a potential molecular diagnostic marker for breast cancer. Tumor Biol. 2016;37:7565–72.
https://doi.org/10.1007/s13277-015-4624-3
Leszinski G, Lehner J, Gezer U, Holdenrieder S. Increased DNA integrity in colorectal cancer. In Vivo. 2014;28(3):299–303. PubMed
Sun Y, An K, Yang C. Circulating cell-free DNA. In: Strumfa I, Gardovskis J, editors. Liquid Biopsy. IntechOpen; 2019.
https://doi.org/10.5772/intechopen.80730
Yan Y, Guo Q, Wang F, Adhikari R, Zhu Z, Zhang H, et al. Cell-free DNA: Hope and potential application in cancer. Front Cell Dev Biol. 2021;9:639233.
https://doi.org/10.3389/fcell.2021.639233
Grunt M, Hillebrand T, Schwarzenbach H. Clinical relevance of size selection of circulating DNA. Transl Cancer Res. 2018;7:S171–84.
https://doi.org/10.21037/tcr.2017.10.10
Gezer U, Bronkhorst AJ, Holdenrieder S. The utility of repetitive cell-free DNA in cancer liquid biopsies. Diagnostics. 2022;12:1363.
https://doi.org/10.3390/diagnostics12061363
Swarup N, Leung HY, Choi I, Aziz MA, Cheng JC, Wong DTW. Cell-free DNA: Features and attributes shaping the next frontier in liquid biopsy. Mol Diagn Ther. 2025;29:277–90.
https://doi.org/10.1007/s40291-025-00773-x
Bronkhorst AJ, Aucamp J, Pretorius PJ. Cell-free DNA: Preanalytical variables. Clinica Chimica Acta. 2015;450:243–53.
https://doi.org/10.1016/j.cca.2015.08.028
Peng H, Pan M, Zhou Z, Chen C, Xing X, Cheng S, et al. The impact of preanalytical variables on the analysis of cell-free DNA from blood and urine samples. Front Cell Dev Biol. 2024;12:1385041.
https://doi.org/10.3389/fcell.2024.1385041
El Messaoudi S, Rolet F, Mouliere F, Thierry AR. Circulating cell free DNA: Preanalytical considerations. Clinica Chimica Acta. 2013;424:222–30.
https://doi.org/10.1016/j.cca.2013.05.022
Casadio V, Salvi S, editors. Cell-free DNA as diagnostic markers: Methods and protocols. New York, NY: Springer New York; 2019.
https://doi.org/10.1007/978-1-4939-8973-7
Lam NYL, Rainer TH, Chiu RWK, Lo YMD. EDTA is a better anticoagulant than heparin or citrate for delayed blood processing for plasma DNA analysis. Clinical Chemistry. 2004;50:256–7.
https://doi.org/10.1373/clinchem.2003.026013
Barra GB, Santa Rita TH, Vasques JDA, Chianca CF, Nery LFA, Costa SSS. EDTA-mediated inhibition of DNases protects circulating cell-free DNA from ex vivo degradation in blood samples. Clinical Biochemistry. 2015;48:976–81.
https://doi.org/10.1016/j.clinbiochem.2015.02.014
Volckmar A, Sültmann H, Riediger A, Fioretos T, Schirmacher P, Endris V, et al. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosomes & Cancer. 2018;57:123–39.
https://doi.org/10.1002/gcc.22517
Sorber L, Zwaenepoel K, Jacobs J, De Winne K, Goethals S, Reclusa P, et al. Circulating cell-free DNA and RNA analysis as liquid biopsy: Optimal centrifugation protocol. Cancers. 2019;11:458.
https://doi.org/10.3390/cancers11040458
Jung M, Klotzek S, Lewandowski M, Fleischhacker M, Jung K. Changes in concentration of DNA in serum and plasma during storage of blood samples. Clinical Chemistry. 2003;49:1028–9.
https://doi.org/10.1373/49.6.1028
Board RE, Williams VS, Knight L, Shaw J, Greystoke A, Ranson M, et al. Isolation and extraction of circulating tumor DNA from patients with small cell lung cancer. Annals of the New York Academy of Sciences. 2008;1137:98–107.
https://doi.org/10.1196/annals.1448.020
Kang Q, Henry NL, Paoletti C, Jiang H, Vats P, Chinnaiyan AM, et al. Comparative analysis of circulating tumor DNA stability in K3EDTA, Streck, and CellSave blood collection tubes. Clinical Biochemistry. 2016;49:1354–60.
https://doi.org/10.1016/j.clinbiochem.2016.03.012
Markus H, Contente-Cuomo T, Farooq M, Liang WS, Borad MJ, Sivakumar S, et al. Evaluation of pre-analytical factors affecting plasma DNA analysis. Sci Rep. 2018;8(1):7375. PubMed
https://doi.org/10.1038/s41598-018-25810-0
Zhao Y, Li Y, Chen P, Li S, Luo J, Xia H. Performance comparison of blood collection tubes as liquid biopsy storage system for minimizing cfDNA contamination from genomic DNA. J Clin Lab Anal. 2019;33(2):e22670. PubMed
https://doi.org/10.1002/jcla.22670
Gerber T, Taschner-Mandl S, Saloberger-Sindhöringer L, Popitsch N, Heitzer E, Witt V, et al. Assessment of pre-analytical sample handling conditions for comprehensive liquid biopsy analysis. The Journal of Molecular Diagnostics. 2020;22:1070–86.
https://doi.org/10.1016/j.jmoldx.2020.05.006
Nesic M, Bødker JS, Terp SK, Dybkær K. Optimization of preanalytical variables for cfDNA processing and detection of ctDNA in archival plasma samples. BioMed Research International. 2021;2021:5585148.
https://doi.org/10.1155/2021/5585148
Van Paemel R, De Koker A, Caggiano C, Morlion A, Mestdagh P, De Wilde B, et al. Genome-wide study of the effect of blood collection tubes on the cell-free DNA methylome. Epigenetics. 2021;16:797–807.
https://doi.org/10.1080/15592294.2020.1827714
Sozzi G, Roz L, Conte D, Mariani L, Andriani F, Verderio P, et al. Effects of prolonged storage of whole plasma or isolated plasma DNA on the results of circulating DNA quantification assays. JNCI: Journal of the National Cancer Institute. 2005;97:1848–50.
https://doi.org/10.1093/jnci/dji432
Larsen PA, Hunnicutt KE, Larsen RJ, Yoder AD, Saunders AM. Warning SINEs: Alu elements, evolution of the human brain, and the spectrum of neurological disease. Chromosome Res. 2018;26:93–111.
https://doi.org/10.1007/s10577-018-9573-4
Hsueh Y-M, Chen M-C, Lin Y-C, Wu C-Y, Shiue H-S, Hsu S-L, et al. Associations among global long interspersed nuclear element-1 DNA methylation, metal exposure, and chronic kidney disease. Arch Toxicol. 2024;98:3127–35.
https://doi.org/10.1007/s00204-024-03780-9
Cheng J, Tang Q, Cao X, Burwinkel B. Cell-free circulating DNA integrity based on peripheral blood as a biomarker for diagnosis of cancer: A systematic review. Cancer Epidemiology, Biomarkers & Prevention. 2017;26:1595–602.
https://doi.org/10.1158/1055-9965.EPI-17-0502
Sobhani N, Generali D, Zanconati F, Bortul M, Scaggiante B. Cell-free DNA integrity for the monitoring of breast cancer: Future perspectives? WJCO. 2018;9:26–32.
https://doi.org/10.5306/wjco.v9.i2.26
Wang X, Shi X-Q, Zeng P-W, Mo F-M, Chen Z-H. Circulating cell free DNA as the diagnostic marker for colorectal cancer: A systematic review and meta-analysis. Oncotarget. 2018;9:24514–24.
https://doi.org/10.18632/oncotarget.25314
Gianni C, Palleschi M, Merloni F, Di Menna G, Sirico M, Sarti S, et al. Cell-free DNA fragmentomics: A promising biomarker for diagnosis, prognosis and prediction of response in breast cancer. IJMS. 2022;23:14197.
https://doi.org/10.3390/ijms232214197
Shaban S, Al-Rahim A, Suleiman A. ALU repeat as potential molecular marker in the detection and prognosis of different cancer types: A systematic review. Mol Clin Oncol. 2022;16:86.
https://doi.org/10.3892/mco.2022.2519
Rodríguez-Ces AM, Rapado-González Ó, Salgado-Barreira Á, Santos MA, Aroso C, Vinhas AS, et al. Liquid biopsies based on cell-free DNA integrity as a biomarker for cancer diagnosis: A meta-analysis. Diagnostics. 2024;14:1465.
https://doi.org/10.3390/diagnostics14141465
Sobhani N, Tierno D, Pavan N, Generali D, Grassi G, Zanconati F, et al. Circulating cell-free DNA integrity for breast and prostate cancer: What is the landscape for clinical management of the most common cancers in women and men? IJMS. 2025;26:900.
https://doi.org/10.3390/ijms26030900
Umetani N, Kim J, Hiramatsu S, Reber HA, Hines OJ, Bilchik AJ, et al. Increased integrity of free circulating DNA in sera of patients with colorectal or periampullary cancer: Direct quantitative PCR for ALU repeats. Clinical Chemistry. 2006;52:1062–9.
https://doi.org/10.1373/clinchem.2006.068577
Madhavan D, Wallwiener M, Bents K, Zucknick M, Nees J, Schott S, et al. Plasma DNA integrity as a biomarker for primary and metastatic breast cancer and potential marker for early diagnosis. Breast Cancer Res Treat. 2014;146:163–74.
https://doi.org/10.1007/s10549-014-2946-2
Chan KCA, Yeung S-W, Lui W-B, Rainer TH, Lo YMD. Effects of preanalytical factors on the molecular size of cell-free DNA in blood. Clin Chem. 2005;51:781–4.
https://doi.org/10.1373/clinchem.2004.046219
Risberg B, Tsui DWY, Biggs H, Ruiz-Valdepenas Martin De Almagro A, Dawson S-J, Hodgkin C, et al. Effects of collection and processing procedures on plasma circulating cell-free DNA from cancer patients. The Journal of Molecular Diagnostics. 2018;20:883–92.
https://doi.org/10.1016/j.jmoldx.2018.07.005
Fleischhacker M, Schmidt B. Pre-analytical issues in liquid biopsy – where do we stand? Journal of Laboratory Medicine. 2020;44:117–42.
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2026 Lifang Zhao, Chao Ying, Songnian Hu, Xuemin Wang, Qimeng Li, Yanning Cai

This work is licensed under a Creative Commons Attribution 4.0 International License.









