Virtual screening of marine coumarins and xanthenes identifies novel acid-suppressive leads targeting histamine H₂ receptor and gastric proton pump
DOI:
https://doi.org/10.17305/bb.2026.13660Keywords:
Marine compounds, coumarin, xanthene, histamine H₂ receptor, gastric H⁺/K⁺-ATPase, virtual screeningAbstract
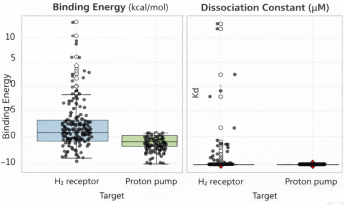
Marine natural products represent a diverse collection of structurally distinct metabolites, many of which have untapped therapeutic potential. This study screened 161 marine-derived coumarin and xanthene compounds for their binding affinity to the histamine H₂ receptor and the gastric H⁺/K⁺-ATPase, the primary regulators of gastric acid secretion. Docking simulations were performed using curated structures of both targets, followed by an evaluation of the compounds for drug-likeness and predicted absorption, distribution, metabolism, and excretion (ADME) properties. Thirty-four compounds demonstrated a stronger predicted affinity for the H₂ receptor than famotidine; however, only three compounds (1, 5, and 150) met all drug-likeness criteria, achieving quantitative estimates of drug-likeness (QED) values exceeding 0.67. Screening against the proton pump yielded 98 hits with higher affinity than soraprazan, with compound 150 being the only candidate to fulfill all medicinal chemistry filters. Interaction analysis indicated that compound 150 binds to the proton pump in a manner that largely overlaps with soraprazan. Density functional theory (DFT) calculations were utilized to characterize the electronic properties of the most promising compounds. ADME predictions suggested favorable permeability and a low risk for human ether-à-go-go-related gene (hERG) inhibition, although high plasma protein binding and the potential for cytochrome P450 (CYP) inhibition may require further optimization. These findings underscore the potential of pyranocoumarin compound 150, along with xanthene derivatives 1 and 5, as promising candidates for the development of new acid-suppressive agents.
Citations
Downloads
References
Feldman M, Friedman LS, Brandt LJ, editors. Sleisenger and Fordtran's Gastrointestinal and Liver Disease: Pathophysiology, Diagnosis, Management. 11th ed. 2 vols. Philadelphia: Elsevier; 2020.
Katz PO, Dunbar KB, Schnoll-Sussman FH, Greer KB, Yadlapati R, Spechler SJ. ACG clinical guideline for the diagnosis and management of gastroesophageal reflux disease. Am J Gastroenterol. 2022;117(1):27–56.
https://doi.org/10.14309/ajg.0000000000001538
Herndiana Y. Functional food in relation to gastroesophageal reflux disease (GERD). Nutrients. 2023;15(16):3583.
https://doi.org/10.3390/nu15163583
Pesce M, Cargiolli M, Cassarano S, Polese B, De Conno B, Aurino L, et al. Diet and functional dyspepsia: clinical correlates and therapeutic perspectives. World J Gastroenterol. 2020;26(5):456–465.
https://doi.org/10.3748/wjg.v26.i5.456
Rang HP, Dale MM, Ritter JM, Flower RJ, Henderson G. Rang & Dale's Pharmacology. 9th ed. Elsevier; 2020.
Tricco AC, Holbrook AM. Histamine H2 receptor antagonists for decreasing gastrointestinal harms in adults using acetylsalicylic acid: systematic review and meta-analysis. Open Med. 2012;6(3):e109–17.
McRorie JW Jr. Rapid development of tachyphylaxis with repeat dosing of H₂-receptor antagonists. Ther Adv Gastroenterol. 2014;5(2):57–62.
https://doi.org/10.4292/wjgpt.v5.i2.57
Aldawsari FS, Alshehry YM, Alghamdi TS. N-nitrosodimethylamine (NDMA) contamination of ranitidine products: a review of recent findings. Saudi Pharm J. 2021;29(1):39–45.
https://doi.org/10.38212/2224-6614.1133
Ketchem CJ, Lynch KL. Potassium-competitive acid blockers and proton pump inhibitors: the dynamic duo of acid blockers. Gastroenterol Hepatol (N Y). 2024;20(12):733–738.
Mori H, Suzuki H. Role of acid suppression in acid-related diseases: proton pump inhibitor and potassium-competitive acid blocker. J Neurogastroenterol Motil. 2019;25(1):6–14.
https://doi.org/10.5056/jnm18139
Tariq R, Singh S, Gupta A, Pardi DS, Khanna S. Association of gastric acid suppression with recurrent Clostridium difficile infection: systematic review and meta-analysis. JAMA Intern Med. 2017;177(6):784–791.
https://doi.org/10.1001/jamainternmed.2017.0212
Klatte DCF, Gasparini A, Xu H, de Deco P, Trevisan M, Johansson ALV, et al. Proton pump inhibitors and risk of chronic kidney disease: a systematic review and meta-analysis. Gastroenterology. 2017;153(3):702–710.
https://doi.org/10.1053/j.gastro.2017.05.046
Ochoa D, Román M, Cabaleiro T, Saiz-Rodríguez M, Mejía G, Abad-Santos F. Effect of food on the pharmacokinetics of omeprazole, pantoprazole and rabeprazole. BMC Pharmacol Toxicol. 2020;21:16.
https://doi.org/10.1186/s40360-020-00433-2
Antequera CM, Orleck K, Jacob R, Kenneally A, Wright WL. Potassium-competitive acid blockers: rethinking acid suppression for gastroesophageal reflux disease and Helicobacter pylori. Postgrad Med. 2024;136(2):131–140.
https://doi.org/10.1080/00325481.2024.2320081
Annunziata F, Pinna C, Dallavalle S, Tamborini L, Pinto A. An overview of coumarin as a versatile and readily accessible scaffold with broad-ranging biological activities. Int J Mol Sci. 2020;21(13):4618.
https://doi.org/10.3390/ijms21134618
Gomes SA, Brandão P, Garcia Fernandes CS, Ramos Pinto Correia da Silva M, Emília DSPDS, de Magalhães Pinto M. Drug-like properties and ADME of xanthone derivatives: the antechamber of clinical trials. Curr Med Chem. 2016;23(32):3654–3686.
https://doi.org/10.2174/0929867323666160425113058
Bouhaoui A, Eddahmi M, Dib M, Khouili M, Aires A, Catto M, et al. Synthesis and biological properties of coumarin derivatives: a review. ChemistrySelect. 2021;6(24):5848–5870.
https://doi.org/10.1002/slct.202101346
Maia M, Resende DI, Duraes F, Pinto MM, Sousa E. Xanthenes in medicinal chemistry—synthetic strategies and biological activities. Eur J Med Chem. 2021;210:113085.
https://doi.org/10.1016/j.ejmech.2020.113085
Gallorini M, Carradori S, Resende DISP, Saso L, Ricci A, Palmeira A, et al. Natural and synthetic xanthone derivatives counteract oxidative stress via Nrf2 modulation in inflamed human macrophages. Int J Mol Sci. 2022;23(21):13319.
https://doi.org/10.3390/ijms232113319
Bai M, Zheng CJ, Nong XH, Zhou XM, Luo YP, Chen GY. Four new insecticidal xanthene derivatives from the mangrove-derived fungus Penicillium sp. JY246. Mar Drugs. 2019;17(12):649.
https://doi.org/10.3390/md17120649
Cao GP, Xia JL, Zhao LY, Tang ZZ, Lin X, Liu YH, et al. Penicixanthene E, a new xanthene isolated from a mangrove-derived fungus Penicillium sp. J Antibiot. 2022;75(9):526–529.
https://doi.org/10.1038/s41429-022-00548-0
Song F, Lin R, Yang N, Jia J, Wei S, Han J, et al. Antibacterial secondary metabolites from marine-derived Aspergillus sp. IMCASMF180035. Antibiotics. 2021;10(4):377.
https://doi.org/10.3390/antibiotics10040377
Blunt JW, Carroll AR, Copp BR, Davis RA, Keyzers RA, Prinsep MR. Marine natural products. Nat Prod Rep. 2016;34(1):235–294.
https://doi.org/10.1039/C7NP00052A
Soares JX, Loureiro DR, Dias AL, Reis S, Pinto MM, Afonso CM. Bioactive marine xanthones: a review. Mar Drugs. 2022;20(1):58.
https://doi.org/10.3390/md20010058
Fernández-Peña L, João Matos M, López E. Recent advances in biologically active coumarins from marine sources: synthesis and evaluation. Mar Drugs. 2022;21(1):37.
https://doi.org/10.3390/md21010037
Veríssimo ACS, Pinto DCGA, Silva AMS. Marine-derived xanthones from 2010 to 2021: isolation, biosynthesis, and biological activities. Mar Drugs. 2022;20(6):347.
https://doi.org/10.3390/md20060347
Reyes-Chilpa R, Baggio CH, Alavez-Solano D, Estrada-Muñiz E, Kauffman FC, Sanchez RI, et al. Inhibition of gastric H⁺,K⁺-ATPase activity by flavonoids, coumarins and xanthones isolated from Mexican medicinal plants. J Ethnopharmacol. 2006;105(1–2):167–172.
https://doi.org/10.1016/j.jep.2005.10.014
Shahzadi K, Bukhari SM, Zaidi A, Wani TA, Jan MS, Zargar S, et al. Novel coumarin derivatives as potential urease inhibitors for kidney stone prevention and anti-ulcer therapy: from synthesis to in vivo evaluation. Pharmaceuticals (Basel). 2023;16(11):1552.
https://doi.org/10.3390/ph16111552
Padole SS, Asnani AJ, Chaple DR, Katre SG. A review of approaches in computer-aided drug design in drug discovery. GSC Biol Pharm Sci. 2022;19(2):075–083.
https://doi.org/10.30574/gscbps.2022.19.2.0161
Pei Z. Computer-aided drug discovery: from traditional simulation methods to language models and quantum computing. Cell Rep Phys Sci. 2024;5:102334.
https://doi.org/10.1016/j.xcrp.2024.102334
Agu PC, Afiukwa CA, Orji OU, Ezeh EM, Ofoke IH, Ogbu CO, et al. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in disease management. Sci Rep. 2023;13(1):13398.
https://doi.org/10.1038/s41598-023-40160-2
Bickerton GR, Paolini GV, Besnard J, Muresan S, Hopkins AL. Quantifying the chemical beauty of drugs. Nat Chem. 2012;4(2):90–98.
https://doi.org/10.1038/nchem.1243
Vazquez-Rodriguez S, Matos MJ, Borges F, Uriarte E, Santana L. Bioactive coumarins from marine sources: origin, structural features and pharmacological properties. Curr Top Med Chem. 2015;15(17):1755–1766.
https://doi.org/10.2174/1568026615666150427125916
Guerriero A, D'Ambrosio M, Cuomo V, Pietra F. A novel, degraded polyketidic lactone, leptosphaerolide, and its likely diketone precursor, leptosphaerodione. Isolation from cultures of the marine ascomycete Leptosphaeria oraemaris (Linder). Helv Chim Acta. 1991;74(7):1445–1450.
https://doi.org/10.1002/hlca.19910740707
Chakraborty K, Antony T, Joy M. Prospective natural anti-inflammatory drimanes attenuating pro-inflammatory 5-lipoxygenase from marine macroalga Gracilaria salicornia. Algal Res. 2019;40:101472.
https://doi.org/10.1016/j.algal.2019.101472
Eltamany EE, Abdelmohsen UR, Ibrahim AK, Hassanean HA, Hentschel U, Ahmed SA. New antibacterial xanthone from the marine sponge-derived Micrococcus sp. EG45. Bioorg Med Chem Lett. 2014;24(21):4939–4942.
https://doi.org/10.1016/j.bmcl.2014.09.040
Fredimoses M, Zhou X, Ai W, Tian X, Yang B, Lin X, et al. Emerixanthone E, a new xanthone derivative from deep-sea fungus Emericella sp. SCSIO 05240. Nat Prod Res. 2019;33(14):2088–2094.
https://doi.org/10.1080/14786419.2018.1487966
He KY, Zhang C, Duan YR, Huang GL, Yang CY, Lu XR, et al. New chlorinated xanthone and anthraquinone produced by a mangrove-derived fungus Penicillium citrinum HL-5126. J Antibiot. 2017;70(7):823–827.
https://doi.org/10.1038/ja.2017.52
Hou JR, Wang YH, Zhong YN, Che TT, Hu Y, Bao J, et al. Protective effect of flavonoids from a deep-sea-derived Arthrinium sp. against ox-LDL-induced oxidative injury through activating AKT/Nrf2/HO-1 pathway in vascular endothelial cells. Mar Drugs. 2021;19(12):712.
https://doi.org/10.3390/md19120712
Kang HH, Zhang HB, Zhong MJ, Ma LY, Liu DS, Liu WZ, et al. Potential antiviral xanthones from a coastal saline soil fungus Aspergillus iizukae. Mar Drugs. 2018;16(11):449.
https://doi.org/10.3390/md16110449
Kong F, Carter GT. Remisporine B, a novel dimeric chromenone derived from spontaneous Diels-Alder reaction of remisporine A. Tetrahedron Lett. 2003;44(15):3119–3122.
https://doi.org/10.1016/S0040-4039(03)00518-5
Li HL, Li XM, Liu H, Meng LH, Wang BG. Two new diphenylketones and a new xanthone from Talaromyces islandicus EN-501, an endophytic fungus. Mar Drugs. 2016;14(12):223.
https://doi.org/10.3390/md14120223
Li X, Li XM, Wang BG. Structural revision of wentiquinone C and related congeners from anthraquinones to xanthones using chemical derivatization and NMR analysis. Mar Drugs. 2018;17(1):8.
https://doi.org/10.3390/md17010008
Liu FA, Lin X, Zhou X, Chen M, Huang X, Yang B, et al. Xanthones and quinolones derivatives produced by the deep-sea-derived fungus Penicillium sp. SCSIO Ind16F01. Molecules. 2017;22(12):1999.
https://doi.org/10.3390/molecules22121999
Ngan NT, Quang TH, Kim KW, Kim HJ, Sohn JH, Kang DG, et al. Anti-inflammatory effects of secondary metabolites from marine-derived Penicillium sp. strain SF-5629. Arch Pharm Res. 2017;40:328–337.
https://doi.org/10.1007/s12272-017-0890-5
Pan JH, Deng JJ, Chen YG, Gao JP, Lin YC, She ZG, et al. New lactone and xanthone derivatives produced by a mangrove endophytic fungus Phoma sp. SK3RW1M. Helv Chim Acta. 2010;93(7):1369–1374.
https://doi.org/10.1002/hlca.200900396
Resende DI, Almeida JR, Pereira S, Campos A, Lemos A, Plowman JE, et al. From natural xanthones to synthetic C-1 aminated 3,4-dioxygenated xanthones as optimized antifouling agents. Mar Drugs. 2021;19(11):638.
https://doi.org/10.3390/md19110638
Shao C, Wang C, Wei M, Gu Y, Xia X, She Z, et al. Two new xanthone derivatives from the marine fungus Penicillium sp. (ZZF 32#). Magn Reson Chem. 2008;46(11):1066–1069.
https://doi.org/10.1002/mrc.2293
Wang CN, Lu HM, Gao CH, Guo L, Zhan ZY, Wang JJ, et al. Cytotoxic benzopyranone and xanthone derivatives from coral symbiotic fungus Cladosporium halotolerans GXIMD 02502. Nat Prod Res. 2021;35(24):5596–5603.
https://doi.org/10.1080/14786419.2020.1799363
Wang HJ, Gloer JB, Scott JA, Malloch D. Coniochaetones A and B: new antifungal benzopyranones from Coniochaeta saccardoi. Tetrahedron Lett. 1995;36(33):5847–5850.
https://doi.org/10.1016/0040-4039(95)01174-G
Wang J, Ding W, Wang R, Du Y, Liu H, Kong X, et al. Identification and bioactivity of compounds from mangrove endophytic fungus Alternaria sp. Mar Drugs. 2015;13(7):4492–4504.
https://doi.org/10.3390/md13074492
Wang P, Luo YF, Zhang M, Dai JG, Wang WJ, Wu J. Three xanthone dimers from Thai mangrove endophytic fungus Phomopsis sp. xy21. J Asian Nat Prod Res. 2018;20(3):217–226.
https://doi.org/10.1080/10286020.2017.1333497
Wang X, Mao ZG, Song BB, Chen CH, Xiao WW, Hu B, et al. Structures and bioactivities of metabolites from mangrove-derived fungi in the South China Sea. Mar Drugs. 2013;11(10):3601–3616.
https://doi.org/10.3390/md11103601
Wu G, Yu G, Kurtán T, Mándi A, Peng J, Mo X, et al. Versixanthones A–F, cytotoxic xanthone-chromanone dimers from marine fungus Aspergillus versicolor HDN1009. J Nat Prod. 2015;78(11):2691–2698.
https://doi.org/10.1021/acs.jnatprod.5b00636
Xia MW, Cui CB, Li CW, Wu CJ, Peng JX, Li DH. Rare chromones from marine-derived Penicillium purpurogenum G59. Mar Drugs. 2015;13(8):5219–5236.
https://doi.org/10.3390/md13085219
Yu G, Wu G, Sun Z, Zhang X, Che Q, Gu Q, et al. Cytotoxic tetrahydroxanthone dimers from mangrove-associated Aspergillus versicolor HDN1009. Mar Drugs. 2018;16(9):335.
https://doi.org/10.3390/md16090335
Zhen X, Gong T, Wen YH, Yan DJ, Chen JJ, Zhu P. Chrysoxanthones A–C: three new xanthone-chromanone heterodimers from sponge-associated Penicillium chrysogenum HLS111. Mar Drugs. 2018;16(10):357.
https://doi.org/10.3390/md16100357
Zhu A, Zhang XW, Zhang M, Li W, Ma ZY, Zhu HJ, et al. Aspergixanthones I–K, new anti-Vibrio prenylxanthones from marine-derived Aspergillus sp. ZA-01. Mar Drugs. 2018;16(9):312.
https://doi.org/10.3390/md16090312
Krieger E, Vriend G. YASARA View—molecular graphics for all devices—from smartphones to workstations. Bioinformatics. 2014;30(20):2981–2982.
https://doi.org/10.1093/bioinformatics/btu426
Krieger E, Vriend G. New ways to boost molecular dynamics simulations. J Comput Chem. 2015;36(13):996–1007.
https://doi.org/10.1002/jcc.23899
Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters. J Chem Theory Comput. 2015;11(8):3696–3713.
https://doi.org/10.1021/acs.jctc.5b00255
Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(1):12–21.
https://doi.org/10.1107/S0907444909042073
Lovell SC, Davis IW, Arendall III WB, De Bakker PI, Word JM, Prisant MG, et al. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins. 2003;50(3):437–450.
https://doi.org/10.1002/prot.10286
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791.
https://doi.org/10.1002/jcc.21256
Bento AP, Hersey A, Félix E, Landrum G, Gaulton A, Atkinson F, et al. An open-source chemical structure curation pipeline using RDKit. J Cheminform. 2020;12(1):51.
https://doi.org/10.1186/s13321-020-00456-1
Shao Y, Molnar LF, Jung Y, Kussmann J, Ochsenfeld C, Brown ST, et al. Advances in methods and algorithms in a modern quantum chemistry program package. Phys Chem Chem Phys. 2006;8(27):3172–3191.
https://doi.org/10.1039/B517914A
Parr RG, Szentpály LV, Liu S. Electrophilicity index. J Am Chem Soc. 1999;121(9):1922–1924.
https://doi.org/10.1021/ja983494x
Xiong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, et al. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49(W1):W5–14.
https://doi.org/10.1093/nar/gkab255
Copeland RA. Evaluation of Enzyme Inhibitors in Drug Discovery: A Guide for Medicinal Chemists and Pharmacologists. 2nd ed. Hoboken (NJ): Wiley; 2013.
https://doi.org/10.1002/9781118540398
Leeson PD, Bento AP, Gaulton A, Hersey A, Manners EJ, Radoux CJ, et al. Target-based evaluation of “drug-like” properties and ligand efficiencies. J Med Chem. 2021;64(11):7210–7230.
https://doi.org/10.1021/acs.jmedchem.1c00416
Robertson MJ, Papasergi-Scott MM, He F, Seven AB, Meyerowitz JG, Panova O, et al. Structure determination of inactive-state GPCRs with a universal nanobody. Nat Struct Mol Biol. 2022;29(12):1188–1195.
https://doi.org/10.1038/s41594-022-00859-8
Tanaka S, Morita M, Yamagishi T, Madapally HV, Hayashida K, Khandelia H, et al. Structural basis for binding of potassium-competitive acid blockers to the gastric proton pump. J Med Chem. 2022;65(11):7843–7853.
https://doi.org/10.1021/acs.jmedchem.2c00338
Leowattana W, Leowattana T. Potassium-competitive acid blockers and gastroesophageal reflux disease. World J Gastroenterol. 2022;28(28):3608–3619.
https://doi.org/10.3748/wjg.v28.i28.3608
Nowak A, Kutyła M, Kaczmarek J, Jaroszuk-Ściseł J, Jędryczka M. Differences in the production of extracellular polymeric substances (EPS) and other metabolites of Plenodomus (Leptosphaeria) infecting winter oilseed rape (Brassica napus L.). Metabolites. 2023;13(6):759.
https://doi.org/10.3390/metabo13060759
Ballante F, Kooistra AJ, Kampen S, de Graaf C, Carlsson J. Structure-based virtual screening for ligands of G protein-coupled receptors: what can molecular docking do for you? Pharmacol Rev. 2021;73(4):1698–1736.
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2026 Amar Osmanović, Mirsada Salihovic, Amila Mehmedović, Amila Turalić, Elma Veljović, Mirha Pazalja, Simone Carradori, Selma Špirtović-Halilović

This work is licensed under a Creative Commons Attribution 4.0 International License.









